Probe CUST_52766_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52766_PI426222305 | JHI_St_60k_v1 | DMT400082826 | ATTCTTTGGCAAAACTCTTCGTGATTTGGTTCATCTGGTTGAAAGTTATGTCTTTCACAC |
All Microarray Probes Designed to Gene DMG400032839
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52766_PI426222305 | JHI_St_60k_v1 | DMT400082826 | ATTCTTTGGCAAAACTCTTCGTGATTTGGTTCATCTGGTTGAAAGTTATGTCTTTCACAC |
| CUST_52765_PI426222305 | JHI_St_60k_v1 | DMT400082827 | GTGCATCAGAGTGTTTGATGTTATGTCTGAGTCATTTTATATGGACTATCTGCTTCTTTT |
Microarray Signals from CUST_52766_PI426222305
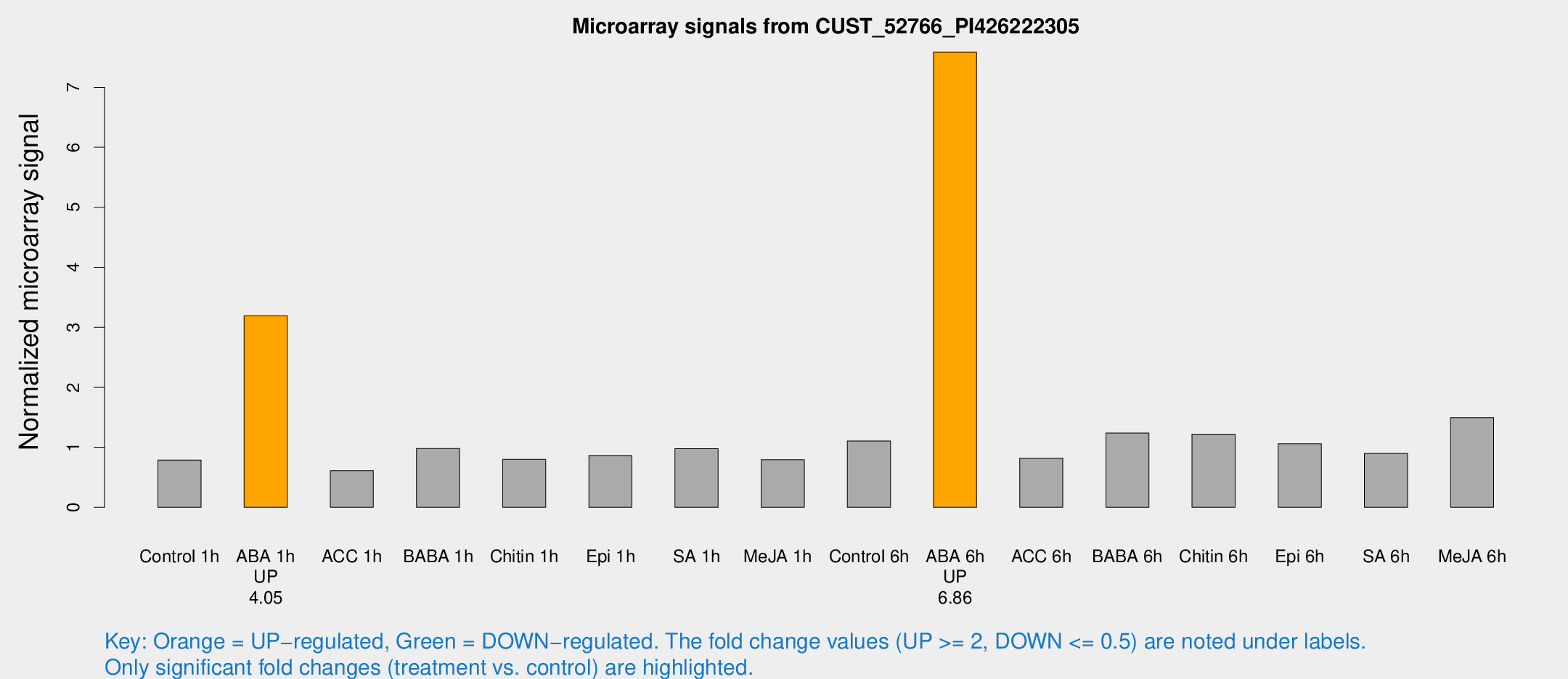
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 423.13 | 41.8009 | 0.788142 | 0.0581882 |
| ABA 1h | 1536.11 | 215.998 | 3.1942 | 0.356145 |
| ACC 1h | 348.678 | 67.2824 | 0.611976 | 0.0875158 |
| BABA 1h | 517.784 | 83.8027 | 0.980469 | 0.0727383 |
| Chitin 1h | 388.041 | 49.6375 | 0.79914 | 0.055457 |
| Epi 1h | 410.08 | 69.9354 | 0.863262 | 0.153575 |
| SA 1h | 538.533 | 46.7539 | 0.977876 | 0.0567593 |
| Me-JA 1h | 348.215 | 38.4745 | 0.79287 | 0.046466 |
| Control 6h | 608.128 | 100.799 | 1.10553 | 0.105364 |
| ABA 6h | 4475.62 | 844.947 | 7.58527 | 1.31899 |
| ACC 6h | 503.945 | 39.1148 | 0.821063 | 0.0759875 |
| BABA 6h | 738.171 | 42.9003 | 1.23618 | 0.0716293 |
| Chitin 6h | 691.615 | 40.2199 | 1.21768 | 0.109867 |
| Epi 6h | 637.598 | 46.7404 | 1.05773 | 0.160781 |
| SA 6h | 518.84 | 139.695 | 0.898465 | 0.190903 |
| Me-JA 6h | 821.06 | 154.632 | 1.49432 | 0.163548 |
Source Transcript PGSC0003DMT400082826 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
