Probe CUST_52734_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52734_PI426222305 | JHI_St_60k_v1 | DMT400003250 | ATTGGTGGCATTTTCCCTCAAGGACAATAGTCTTCACAATGAATTCGAGGTCTCCGAGAC |
All Microarray Probes Designed to Gene DMG400001287
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52734_PI426222305 | JHI_St_60k_v1 | DMT400003250 | ATTGGTGGCATTTTCCCTCAAGGACAATAGTCTTCACAATGAATTCGAGGTCTCCGAGAC |
Microarray Signals from CUST_52734_PI426222305
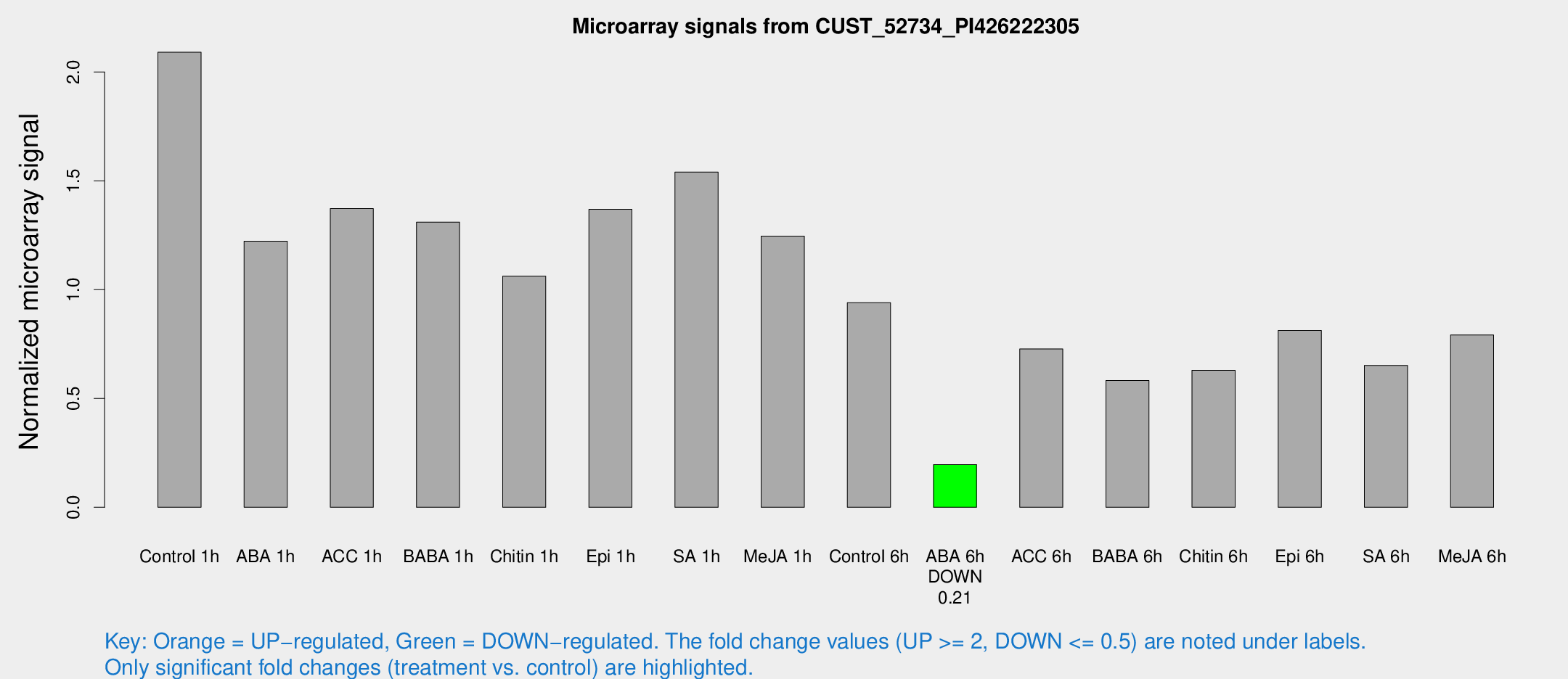
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1580.41 | 166.429 | 2.09102 | 0.129737 |
| ABA 1h | 820.868 | 97.4615 | 1.22248 | 0.143066 |
| ACC 1h | 1285.27 | 457.138 | 1.37235 | 0.648773 |
| BABA 1h | 986.514 | 207.32 | 1.31027 | 0.178712 |
| Chitin 1h | 720.409 | 68.4528 | 1.06206 | 0.0614771 |
| Epi 1h | 893.827 | 84.524 | 1.36907 | 0.138172 |
| SA 1h | 1264.66 | 333.644 | 1.54005 | 0.384932 |
| Me-JA 1h | 766.011 | 62.5107 | 1.24567 | 0.0720914 |
| Control 6h | 784.797 | 226.723 | 0.940487 | 0.257318 |
| ABA 6h | 157.76 | 15.8631 | 0.196295 | 0.0157489 |
| ACC 6h | 632.106 | 68.5135 | 0.727377 | 0.0973658 |
| BABA 6h | 520.301 | 134.342 | 0.582906 | 0.158929 |
| Chitin 6h | 502.161 | 29.2458 | 0.629571 | 0.0366019 |
| Epi 6h | 707.574 | 125.264 | 0.813073 | 0.183436 |
| SA 6h | 499.552 | 87.3335 | 0.6523 | 0.0506074 |
| Me-JA 6h | 609.567 | 111.857 | 0.791589 | 0.0793272 |
Source Transcript PGSC0003DMT400003250 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G02000.1 | +3 | 1e-70 | 226 | 118/151 (78%) | glutamate decarboxylase 3 | chr2:469505-471997 REVERSE LENGTH=500 |
