Probe CUST_52731_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52731_PI426222305 | JHI_St_60k_v1 | DMT400083282 | AGATTTCTGCCGACATTGTTGGAATTTGCTTCCTCTTTTCATGATTTTACAGCTGTTAAT |
All Microarray Probes Designed to Gene DMG401033165
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52731_PI426222305 | JHI_St_60k_v1 | DMT400083282 | AGATTTCTGCCGACATTGTTGGAATTTGCTTCCTCTTTTCATGATTTTACAGCTGTTAAT |
| CUST_52729_PI426222305 | JHI_St_60k_v1 | DMT400083284 | CAAATCATGGAAGATGTTGCATTGGAGGTTAAAGAGAAAATCTTGCATCAGAGGAACAGA |
| CUST_52730_PI426222305 | JHI_St_60k_v1 | DMT400083285 | TTTCATGGATTTGCTGTGACATGCAACAAATCACGGAAGATCTTGCATCAGAGGAACAGA |
Microarray Signals from CUST_52731_PI426222305
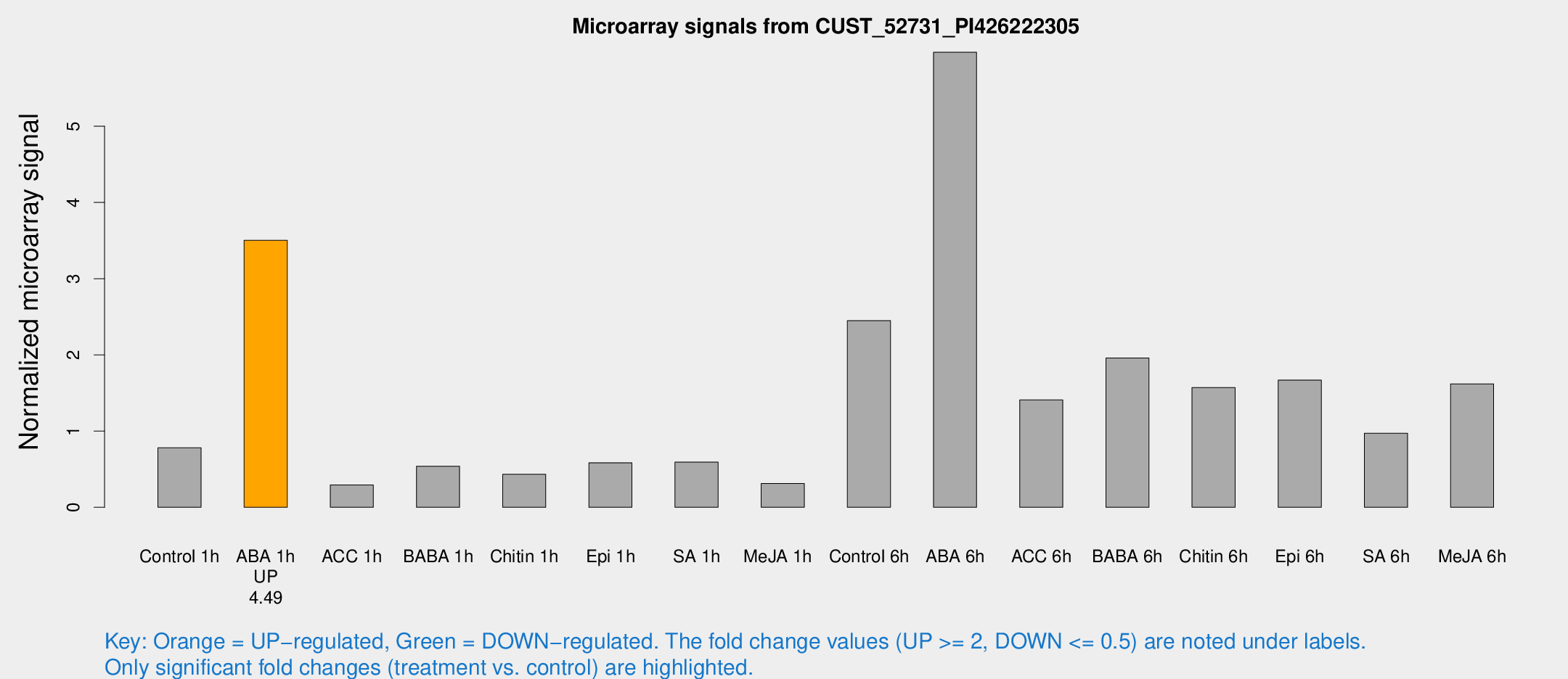
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 39.5068 | 4.1773 | 0.780965 | 0.0833058 |
| ABA 1h | 158.882 | 20.9061 | 3.50353 | 0.3662 |
| ACC 1h | 17.6423 | 6.32272 | 0.292459 | 0.110537 |
| BABA 1h | 30.8625 | 10.252 | 0.539292 | 0.201214 |
| Chitin 1h | 19.7561 | 3.74567 | 0.433846 | 0.0841706 |
| Epi 1h | 30.857 | 11.4277 | 0.584125 | 0.314879 |
| SA 1h | 31.2297 | 4.3062 | 0.593095 | 0.076582 |
| Me-JA 1h | 14.9047 | 5.29696 | 0.312346 | 0.13262 |
| Control 6h | 130.29 | 28.9135 | 2.44897 | 0.404673 |
| ABA 6h | 348.275 | 89.324 | 5.97152 | 1.67708 |
| ACC 6h | 83.1154 | 11.6422 | 1.40898 | 0.156994 |
| BABA 6h | 130.895 | 43.3505 | 1.95898 | 0.94027 |
| Chitin 6h | 88.5573 | 18.431 | 1.57187 | 0.372931 |
| Epi 6h | 101.903 | 28.5304 | 1.66959 | 0.510349 |
| SA 6h | 63.9614 | 29.7026 | 0.97188 | 0.940771 |
| Me-JA 6h | 99.4275 | 37.7601 | 1.61988 | 0.747061 |
Source Transcript PGSC0003DMT400083282 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
