Probe CUST_52729_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52729_PI426222305 | JHI_St_60k_v1 | DMT400083284 | CAAATCATGGAAGATGTTGCATTGGAGGTTAAAGAGAAAATCTTGCATCAGAGGAACAGA |
All Microarray Probes Designed to Gene DMG401033165
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52731_PI426222305 | JHI_St_60k_v1 | DMT400083282 | AGATTTCTGCCGACATTGTTGGAATTTGCTTCCTCTTTTCATGATTTTACAGCTGTTAAT |
| CUST_52730_PI426222305 | JHI_St_60k_v1 | DMT400083285 | TTTCATGGATTTGCTGTGACATGCAACAAATCACGGAAGATCTTGCATCAGAGGAACAGA |
| CUST_52729_PI426222305 | JHI_St_60k_v1 | DMT400083284 | CAAATCATGGAAGATGTTGCATTGGAGGTTAAAGAGAAAATCTTGCATCAGAGGAACAGA |
Microarray Signals from CUST_52729_PI426222305
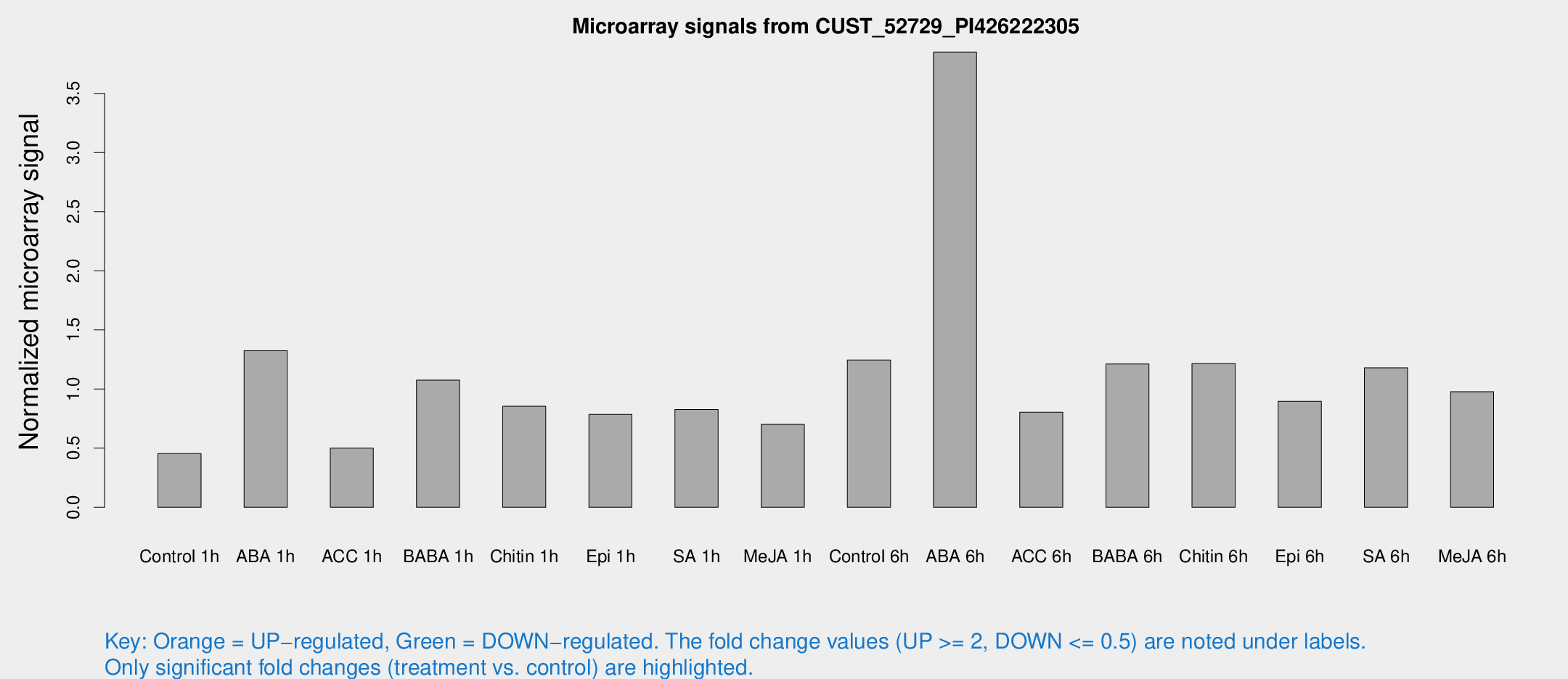
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 45.5589 | 6.83117 | 0.454808 | 0.0427433 |
| ABA 1h | 118.851 | 22.1336 | 1.32386 | 0.226434 |
| ACC 1h | 52.4467 | 9.74357 | 0.500925 | 0.066277 |
| BABA 1h | 108.253 | 25.6073 | 1.07532 | 0.182357 |
| Chitin 1h | 78.2841 | 15.6046 | 0.853985 | 0.156883 |
| Epi 1h | 69.6162 | 13.442 | 0.786108 | 0.167693 |
| SA 1h | 85.6734 | 14.6702 | 0.827301 | 0.0922367 |
| Me-JA 1h | 58.1541 | 11.7476 | 0.700662 | 0.0783253 |
| Control 6h | 125.549 | 20.3352 | 1.24607 | 0.131238 |
| ABA 6h | 416.282 | 84.6402 | 3.84781 | 0.547516 |
| ACC 6h | 97.7075 | 24.2151 | 0.803585 | 0.210522 |
| BABA 6h | 135.432 | 18.2747 | 1.21168 | 0.13583 |
| Chitin 6h | 127.973 | 12.9479 | 1.21554 | 0.116825 |
| Epi 6h | 99.5367 | 7.59611 | 0.896727 | 0.0623299 |
| SA 6h | 117.504 | 18.7127 | 1.17921 | 0.328795 |
| Me-JA 6h | 109.894 | 35.6942 | 0.977256 | 0.342164 |
Source Transcript PGSC0003DMT400083284 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
