Probe CUST_52715_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52715_PI426222305 | JHI_St_60k_v1 | DMT400006338 | GAGGATGATGTTATTACCACATAATAAGAGATGAAGAGTCTCATTTTCTGCTTCTCCTTG |
All Microarray Probes Designed to Gene DMG400002479
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52715_PI426222305 | JHI_St_60k_v1 | DMT400006338 | GAGGATGATGTTATTACCACATAATAAGAGATGAAGAGTCTCATTTTCTGCTTCTCCTTG |
| CUST_52714_PI426222305 | JHI_St_60k_v1 | DMT400006337 | CCCTCTTTTGAGTGACATGTTGATCAATGTTCTTTTTTCTTGGTTGATTGGTCAAATTTC |
Microarray Signals from CUST_52715_PI426222305
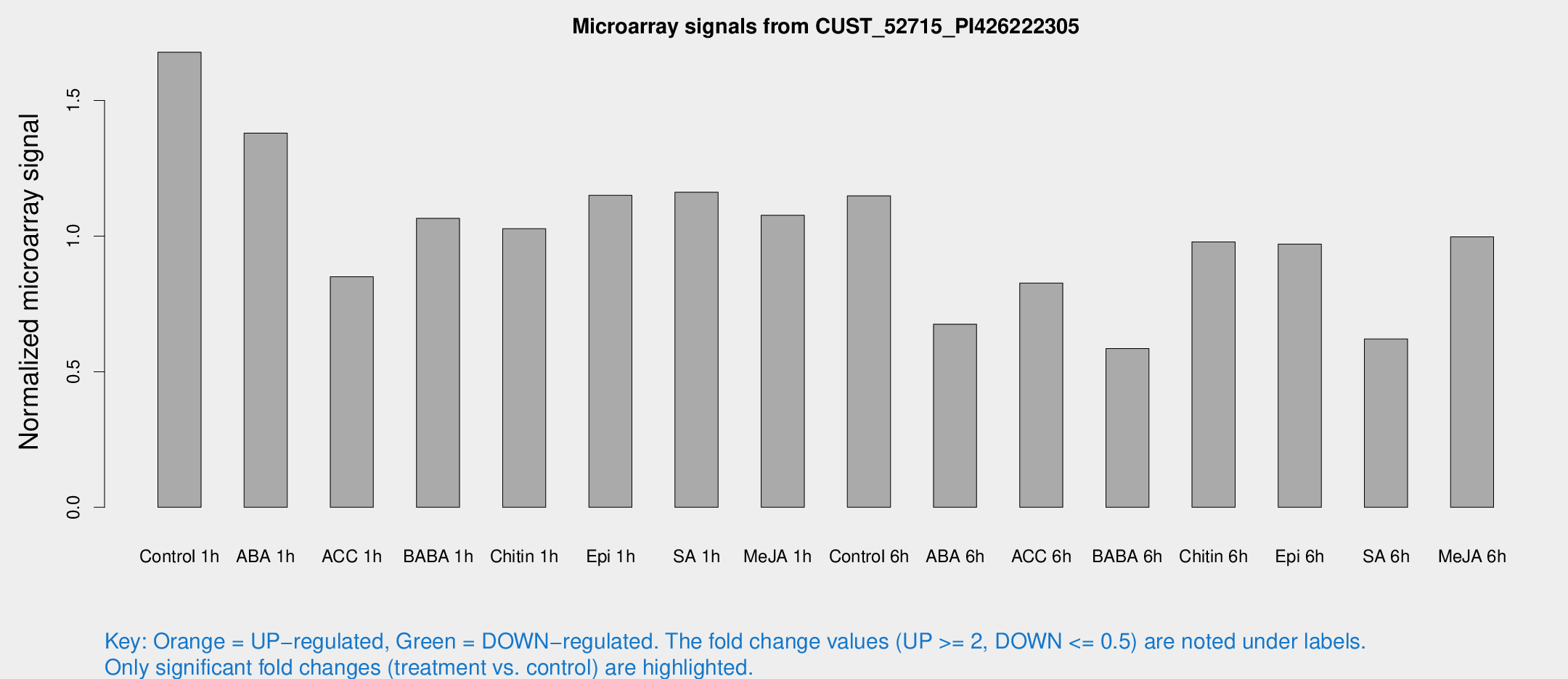
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 28797.5 | 2318.43 | 1.67801 | 0.204847 |
| ABA 1h | 21117.7 | 2622.58 | 1.37974 | 0.171434 |
| ACC 1h | 17687.1 | 6050.54 | 0.849824 | 0.359972 |
| BABA 1h | 18417.1 | 4010.9 | 1.06522 | 0.149315 |
| Chitin 1h | 16364.1 | 3109.05 | 1.02714 | 0.193108 |
| Epi 1h | 17944.6 | 4119.69 | 1.15046 | 0.281236 |
| SA 1h | 20975.7 | 3752.32 | 1.16148 | 0.191263 |
| Me-JA 1h | 15234.3 | 1967.08 | 1.07688 | 0.133874 |
| Control 6h | 20587.4 | 4293.42 | 1.14825 | 0.172017 |
| ABA 6h | 12301.6 | 895.659 | 0.675005 | 0.0389719 |
| ACC 6h | 16425.5 | 2090 | 0.827025 | 0.165206 |
| BABA 6h | 12040.8 | 3101.97 | 0.585553 | 0.189617 |
| Chitin 6h | 17901.1 | 1646.86 | 0.978442 | 0.112118 |
| Epi 6h | 18733.1 | 1199.95 | 0.970747 | 0.0560465 |
| SA 6h | 10885.2 | 1986.75 | 0.620622 | 0.0546882 |
| Me-JA 6h | 18891 | 6265.78 | 0.997511 | 0.316689 |
Source Transcript PGSC0003DMT400006338 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G29320.1 | +1 | 1e-91 | 291 | 140/160 (88%) | Glycosyl transferase, family 35 | chr3:11252871-11257587 FORWARD LENGTH=962 |
