Probe CUST_52714_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52714_PI426222305 | JHI_St_60k_v1 | DMT400006337 | CCCTCTTTTGAGTGACATGTTGATCAATGTTCTTTTTTCTTGGTTGATTGGTCAAATTTC |
All Microarray Probes Designed to Gene DMG400002479
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52714_PI426222305 | JHI_St_60k_v1 | DMT400006337 | CCCTCTTTTGAGTGACATGTTGATCAATGTTCTTTTTTCTTGGTTGATTGGTCAAATTTC |
| CUST_52715_PI426222305 | JHI_St_60k_v1 | DMT400006338 | GAGGATGATGTTATTACCACATAATAAGAGATGAAGAGTCTCATTTTCTGCTTCTCCTTG |
Microarray Signals from CUST_52714_PI426222305
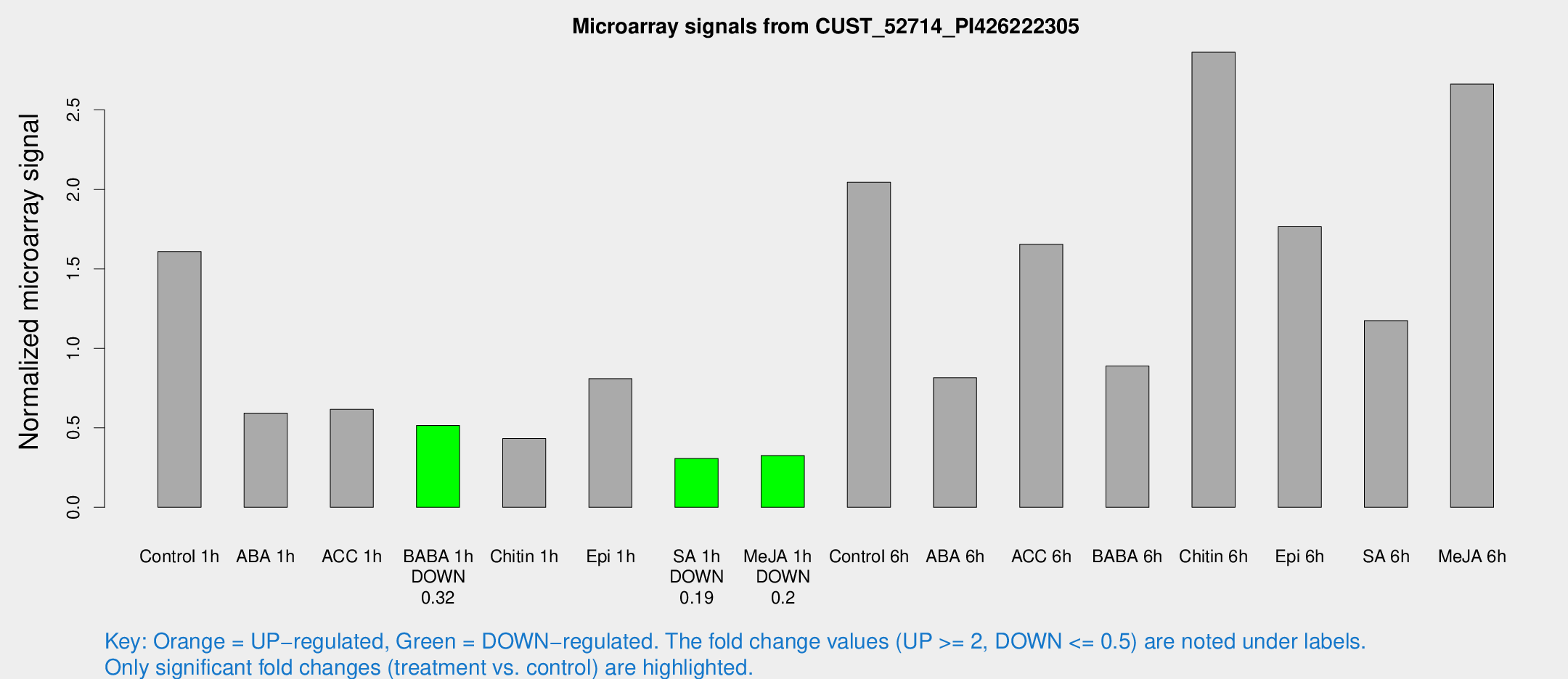
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 34.5569 | 5.75272 | 1.6093 | 0.180584 |
| ABA 1h | 13.3544 | 5.75517 | 0.593019 | 0.291308 |
| ACC 1h | 15.0126 | 4.56113 | 0.616044 | 0.209513 |
| BABA 1h | 11.9186 | 4.3178 | 0.51474 | 0.19864 |
| Chitin 1h | 10.0676 | 4.97864 | 0.432586 | 0.198004 |
| Epi 1h | 15.5762 | 3.68778 | 0.809488 | 0.20981 |
| SA 1h | 6.85385 | 3.13166 | 0.306897 | 0.152118 |
| Me-JA 1h | 5.5807 | 3.25156 | 0.325408 | 0.188689 |
| Control 6h | 48.9362 | 16.9945 | 2.04462 | 0.659515 |
| ABA 6h | 18.7574 | 3.63104 | 0.814808 | 0.211039 |
| ACC 6h | 39.9545 | 4.63633 | 1.65516 | 0.348551 |
| BABA 6h | 24.4072 | 9.60397 | 0.888919 | 0.376241 |
| Chitin 6h | 68.1824 | 18.3726 | 2.86333 | 0.842637 |
| Epi 6h | 43.1855 | 7.67978 | 1.76525 | 0.488114 |
| SA 6h | 31.3461 | 12.2828 | 1.17404 | 1.09236 |
| Me-JA 6h | 71.3475 | 36.1823 | 2.663 | 1.39199 |
Source Transcript PGSC0003DMT400006337 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G29320.1 | +1 | 1e-86 | 276 | 140/193 (73%) | Glycosyl transferase, family 35 | chr3:11252871-11257587 FORWARD LENGTH=962 |
