Probe CUST_52392_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52392_PI426222305 | JHI_St_60k_v1 | DMT400049142 | AAGAAAAAGTATGCACCTATTTGTGTCACAATGCCAACTGCTAGAATATGTCACAAGTGA |
All Microarray Probes Designed to Gene DMG400019098
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52392_PI426222305 | JHI_St_60k_v1 | DMT400049142 | AAGAAAAAGTATGCACCTATTTGTGTCACAATGCCAACTGCTAGAATATGTCACAAGTGA |
Microarray Signals from CUST_52392_PI426222305
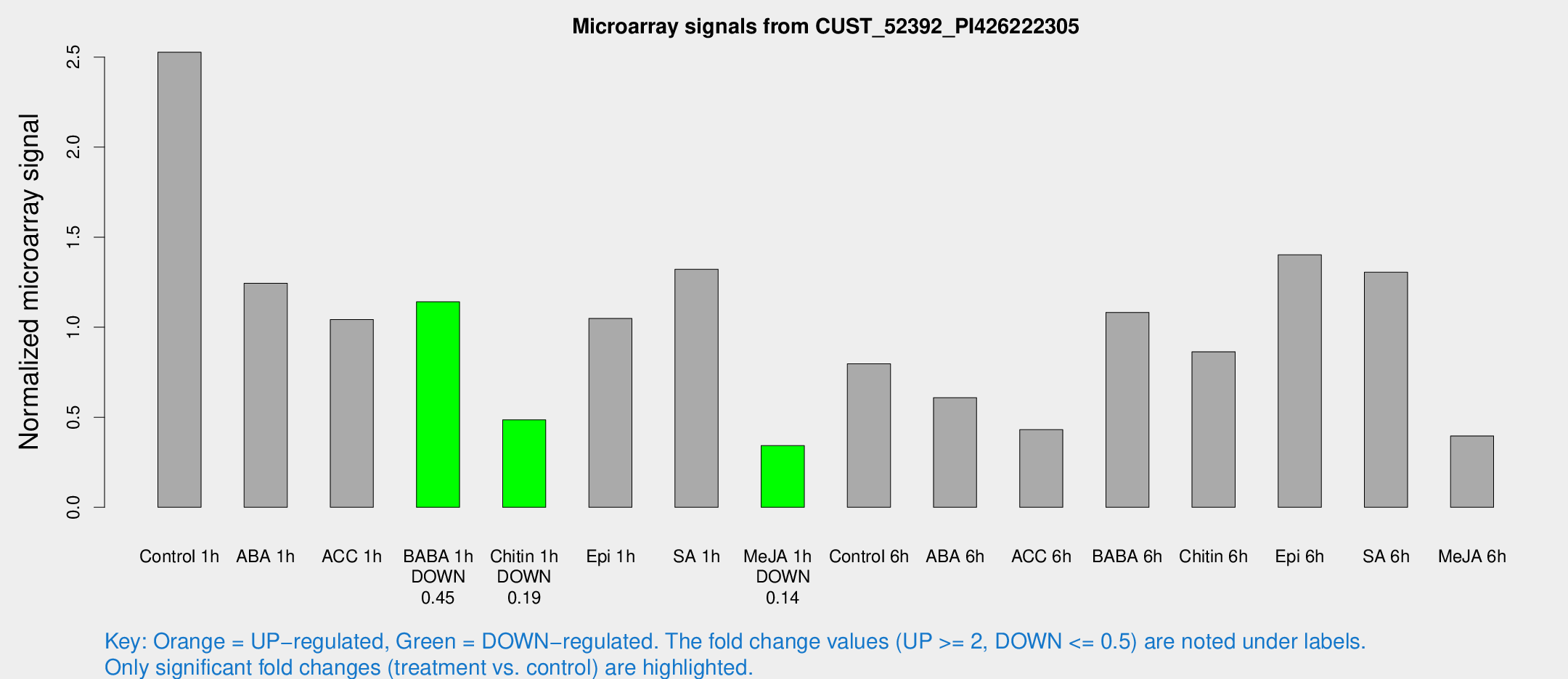
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1626.44 | 286.309 | 2.52692 | 0.268443 |
| ABA 1h | 782.222 | 262.716 | 1.24418 | 0.460537 |
| ACC 1h | 923.24 | 396.46 | 1.04234 | 0.778615 |
| BABA 1h | 739.508 | 179.066 | 1.14087 | 0.211387 |
| Chitin 1h | 329.402 | 139.243 | 0.4857 | 0.220351 |
| Epi 1h | 634.398 | 209.787 | 1.04859 | 0.38284 |
| SA 1h | 913.468 | 261.281 | 1.32195 | 0.289356 |
| Me-JA 1h | 186.148 | 49.4843 | 0.342336 | 0.0623784 |
| Control 6h | 523.472 | 112.024 | 0.796572 | 0.124447 |
| ABA 6h | 419.208 | 75.2801 | 0.608864 | 0.0877301 |
| ACC 6h | 311.261 | 26.3577 | 0.431228 | 0.0702519 |
| BABA 6h | 764.062 | 74.4131 | 1.08228 | 0.0859965 |
| Chitin 6h | 622.955 | 155.949 | 0.863777 | 0.276525 |
| Epi 6h | 1056.13 | 256.959 | 1.40178 | 0.274756 |
| SA 6h | 871.169 | 238.575 | 1.30495 | 0.247374 |
| Me-JA 6h | 349.071 | 206.616 | 0.396218 | 0.241104 |
Source Transcript PGSC0003DMT400049142 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G51400.1 | +1 | 4e-15 | 67 | 47/116 (41%) | Photosystem II 5 kD protein | chr1:19052172-19052492 REVERSE LENGTH=106 |
