Probe CUST_52326_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52326_PI426222305 | JHI_St_60k_v1 | DMT400014262 | TACTGATGACATAGCAACATACGAGGTTGAGAAAGGCAGGGGACAAATTGCAACGGGAAT |
All Microarray Probes Designed to Gene DMG400005595
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52326_PI426222305 | JHI_St_60k_v1 | DMT400014262 | TACTGATGACATAGCAACATACGAGGTTGAGAAAGGCAGGGGACAAATTGCAACGGGAAT |
Microarray Signals from CUST_52326_PI426222305
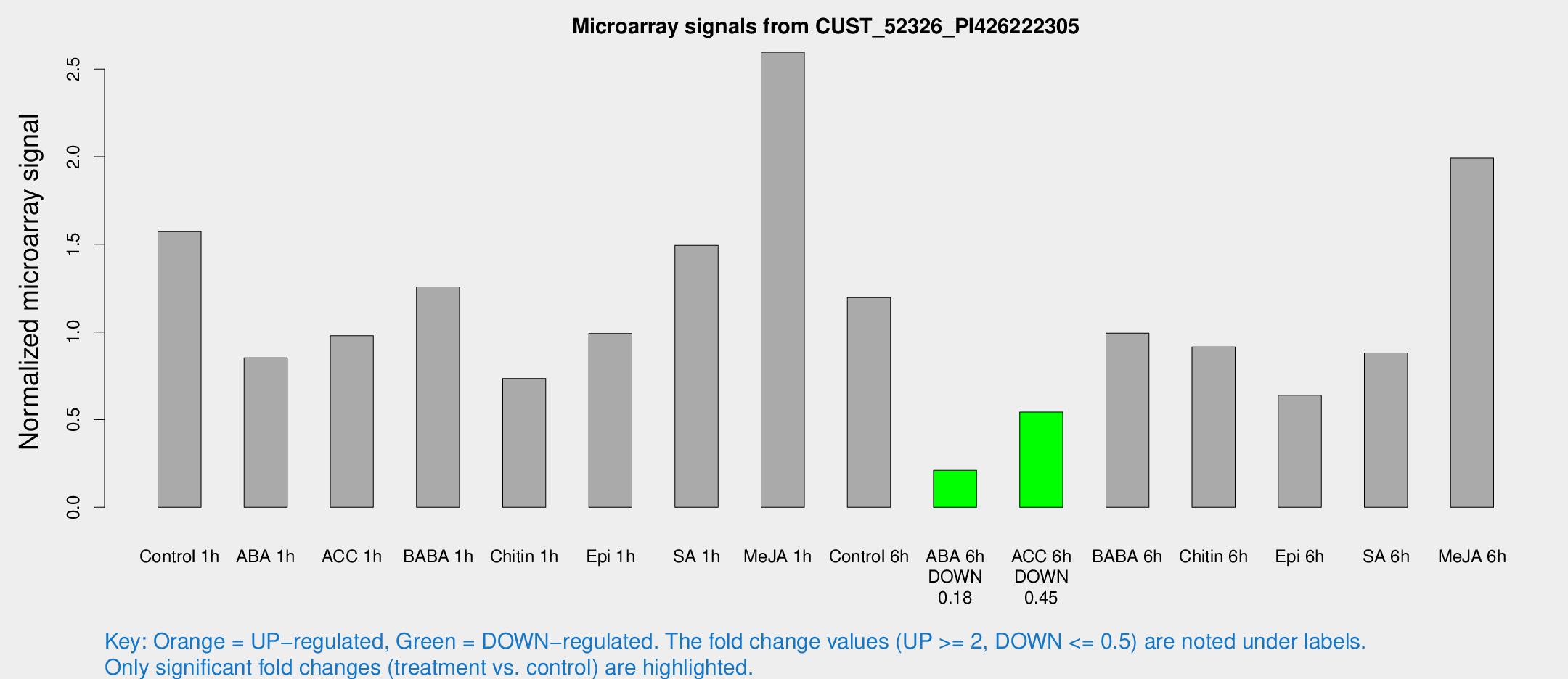
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 589.718 | 129.953 | 1.57255 | 0.260005 |
| ABA 1h | 300.585 | 89.4366 | 0.853131 | 0.244591 |
| ACC 1h | 415.929 | 158.935 | 0.978663 | 0.344635 |
| BABA 1h | 460.66 | 108.8 | 1.25751 | 0.204443 |
| Chitin 1h | 248.538 | 55.4513 | 0.735166 | 0.102763 |
| Epi 1h | 318.493 | 54.8908 | 0.99153 | 0.177409 |
| SA 1h | 558.793 | 69.9878 | 1.49449 | 0.230324 |
| Me-JA 1h | 823.179 | 243.759 | 2.59605 | 0.523479 |
| Control 6h | 452.921 | 103.454 | 1.19624 | 0.20705 |
| ABA 6h | 81.4648 | 8.05321 | 0.211575 | 0.031114 |
| ACC 6h | 226.784 | 27.5942 | 0.543499 | 0.0327298 |
| BABA 6h | 399.544 | 23.4146 | 0.993664 | 0.0832695 |
| Chitin 6h | 355.923 | 50.9338 | 0.914654 | 0.0984657 |
| Epi 6h | 263.879 | 37.1131 | 0.640035 | 0.0754155 |
| SA 6h | 348.838 | 105.724 | 0.880922 | 0.216279 |
| Me-JA 6h | 718.701 | 76.0261 | 1.99161 | 0.218113 |
Source Transcript PGSC0003DMT400014262 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G23960.1 | +1 | 3e-122 | 375 | 218/580 (38%) | terpene synthase 21 | chr5:8092969-8095128 FORWARD LENGTH=547 |
