Probe CUST_52172_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52172_PI426222305 | JHI_St_60k_v1 | DMT400059885 | CATGGTGAAGTAGTGTTGCATGATGATTTGTTTGATAGTGCTAGAATGTGGTCCTTTTGA |
All Microarray Probes Designed to Gene DMG400023295
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52172_PI426222305 | JHI_St_60k_v1 | DMT400059885 | CATGGTGAAGTAGTGTTGCATGATGATTTGTTTGATAGTGCTAGAATGTGGTCCTTTTGA |
Microarray Signals from CUST_52172_PI426222305
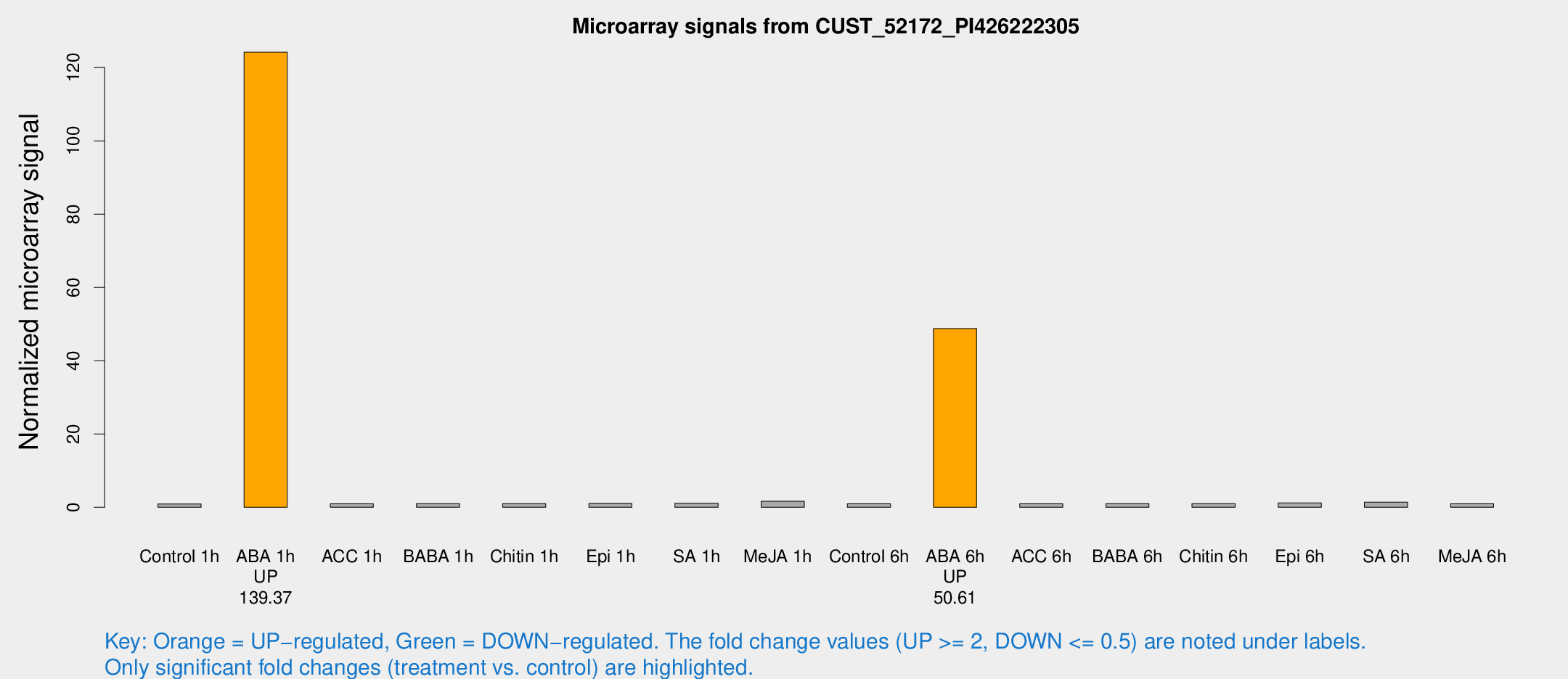
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.38027 | 3.1182 | 0.89101 | 0.516152 |
| ABA 1h | 671.024 | 75.5476 | 124.178 | 15.1689 |
| ACC 1h | 5.91007 | 3.42632 | 0.952644 | 0.551813 |
| BABA 1h | 5.78841 | 3.35544 | 0.993939 | 0.575568 |
| Chitin 1h | 5.53783 | 3.22183 | 1.02414 | 0.588717 |
| Epi 1h | 5.4521 | 3.16595 | 1.04163 | 0.60368 |
| SA 1h | 7.00653 | 3.19036 | 1.08168 | 0.537083 |
| Me-JA 1h | 8.44144 | 3.31379 | 1.64189 | 0.697859 |
| Control 6h | 5.83241 | 3.38274 | 0.963776 | 0.558805 |
| ABA 6h | 339.892 | 88.9301 | 48.7754 | 12.6651 |
| ACC 6h | 6.84299 | 4.05931 | 0.965899 | 0.559324 |
| BABA 6h | 6.72167 | 3.80532 | 0.988924 | 0.560128 |
| Chitin 6h | 6.36268 | 3.68437 | 0.990257 | 0.573385 |
| Epi 6h | 8.22632 | 3.85755 | 1.17998 | 0.583989 |
| SA 6h | 8.54922 | 3.57062 | 1.40065 | 0.619276 |
| Me-JA 6h | 5.86645 | 3.4001 | 0.97522 | 0.564758 |
Source Transcript PGSC0003DMT400059885 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G74930.1 | +1 | 1e-20 | 86 | 56/98 (57%) | Integrase-type DNA-binding superfamily protein | chr1:28144239-28144826 FORWARD LENGTH=195 |
