Probe CUST_52060_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52060_PI426222305 | JHI_St_60k_v1 | DMT400054334 | GCTCATCGATTTTCTCCTGCTGGTGGTTTCCATGTTTACATTAGGTATCGGAAAGGAGCA |
All Microarray Probes Designed to Gene DMG400021087
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_52059_PI426222305 | JHI_St_60k_v1 | DMT400054335 | CCTTTTATGAAACTTTTACCCCATTAATGTTTGGTAAGGCTTGACATTCCTTCTCTGATA |
| CUST_52060_PI426222305 | JHI_St_60k_v1 | DMT400054334 | GCTCATCGATTTTCTCCTGCTGGTGGTTTCCATGTTTACATTAGGTATCGGAAAGGAGCA |
Microarray Signals from CUST_52060_PI426222305
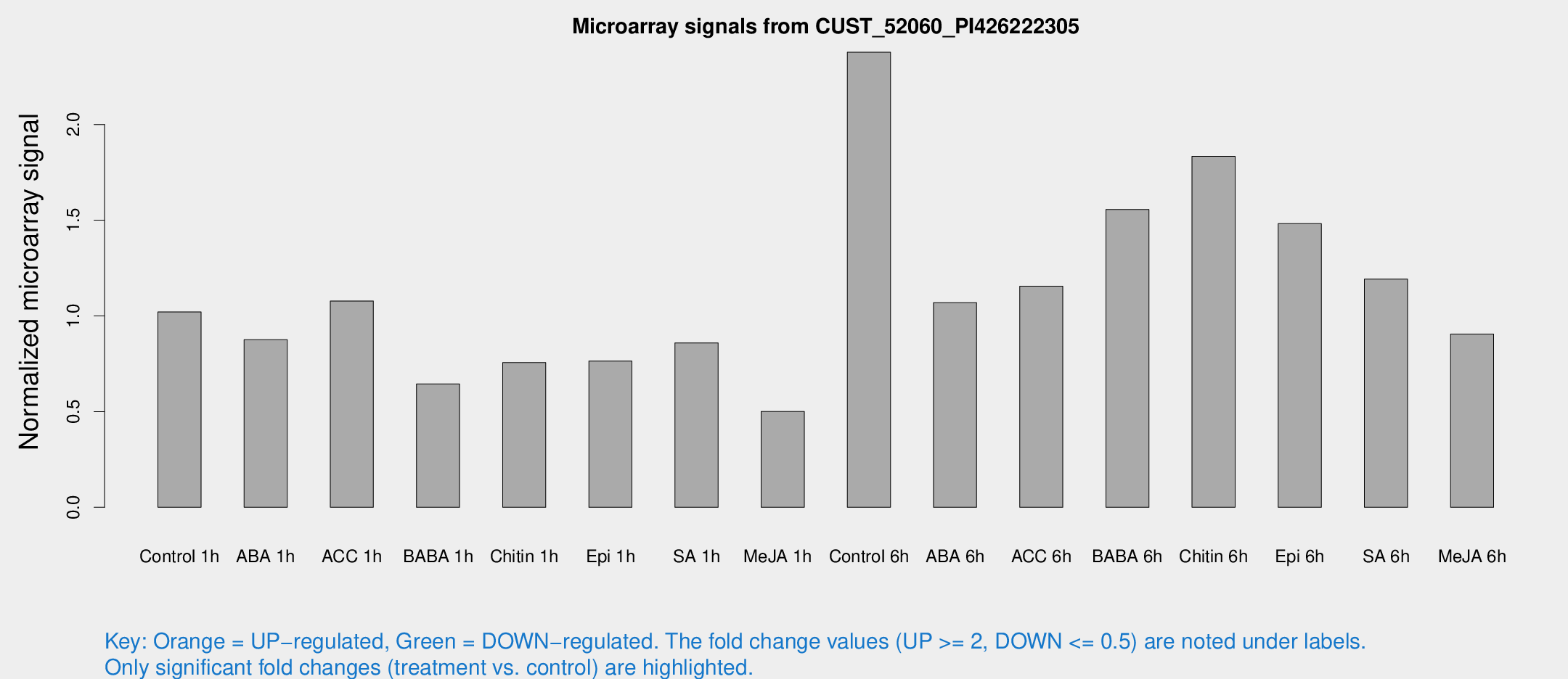
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 76.9878 | 6.85526 | 1.02047 | 0.0728656 |
| ABA 1h | 62.6549 | 18.2199 | 0.875754 | 0.319762 |
| ACC 1h | 87.6999 | 21.7953 | 1.07791 | 0.359241 |
| BABA 1h | 48.6054 | 9.48598 | 0.644466 | 0.0756143 |
| Chitin 1h | 51.6712 | 5.93219 | 0.756453 | 0.151326 |
| Epi 1h | 50.205 | 5.99638 | 0.764326 | 0.0942073 |
| SA 1h | 66.9287 | 7.77762 | 0.85866 | 0.13164 |
| Me-JA 1h | 31.771 | 6.31512 | 0.500339 | 0.0632499 |
| Control 6h | 194.781 | 53.5002 | 2.37776 | 0.560831 |
| ABA 6h | 86.0982 | 8.78235 | 1.06959 | 0.0822344 |
| ACC 6h | 104.97 | 23.6869 | 1.15532 | 0.165819 |
| BABA 6h | 145.251 | 50.1804 | 1.55634 | 0.523574 |
| Chitin 6h | 148.844 | 19.0555 | 1.83413 | 0.269647 |
| Epi 6h | 133.023 | 31.8507 | 1.48192 | 0.468643 |
| SA 6h | 93.8371 | 23.5173 | 1.19269 | 0.201835 |
| Me-JA 6h | 74.375 | 20.0044 | 0.905054 | 0.248974 |
Source Transcript PGSC0003DMT400054334 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G24340.1 | +1 | 2e-101 | 317 | 153/239 (64%) | FAD/NAD(P)-binding oxidoreductase family protein | chr1:8635416-8638866 FORWARD LENGTH=709 |
