Probe CUST_5193_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5193_PI426222305 | JHI_St_60k_v1 | DMT400003777 | ATCCTTCTGTCTACGCTAAATGGGTGTCACTACCTATACACATGAAGCCACAAGCACAGT |
All Microarray Probes Designed to Gene DMG400001495
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5193_PI426222305 | JHI_St_60k_v1 | DMT400003777 | ATCCTTCTGTCTACGCTAAATGGGTGTCACTACCTATACACATGAAGCCACAAGCACAGT |
Microarray Signals from CUST_5193_PI426222305
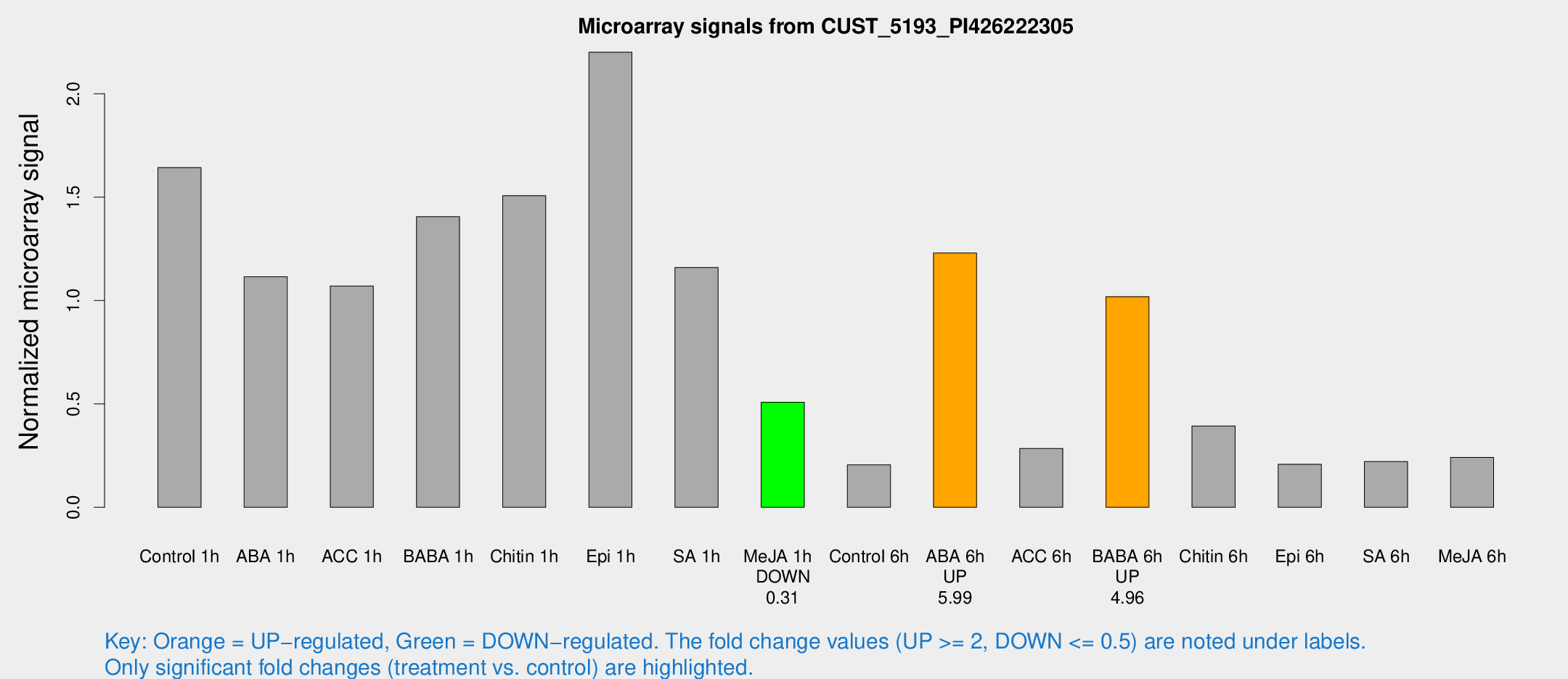
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 41.4303 | 3.78291 | 1.64304 | 0.227098 |
| ABA 1h | 24.7607 | 3.13752 | 1.11486 | 0.141283 |
| ACC 1h | 30.2904 | 8.56685 | 1.07027 | 0.270616 |
| BABA 1h | 35.8748 | 7.57304 | 1.40583 | 0.197451 |
| Chitin 1h | 34.4618 | 3.87455 | 1.50769 | 0.245947 |
| Epi 1h | 49.8064 | 10.3234 | 2.2012 | 0.396372 |
| SA 1h | 30.0944 | 3.40536 | 1.16039 | 0.1328 |
| Me-JA 1h | 10.4567 | 3.02997 | 0.508512 | 0.147573 |
| Control 6h | 5.16729 | 2.99893 | 0.205506 | 0.11925 |
| ABA 6h | 34.7156 | 8.11752 | 1.22996 | 0.280671 |
| ACC 6h | 8.19888 | 3.65637 | 0.284701 | 0.125341 |
| BABA 6h | 28.6334 | 3.73737 | 1.01897 | 0.133263 |
| Chitin 6h | 11.4945 | 3.38312 | 0.3936 | 0.144346 |
| Epi 6h | 5.92832 | 3.47008 | 0.207941 | 0.120774 |
| SA 6h | 5.50191 | 3.17226 | 0.221604 | 0.127745 |
| Me-JA 6h | 6.15369 | 2.98597 | 0.241387 | 0.122329 |
Source Transcript PGSC0003DMT400003777 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G60000.1 | +1 | 2e-86 | 266 | 128/175 (73%) | RNA-binding (RRM/RBD/RNP motifs) family protein | chr1:22093678-22094540 REVERSE LENGTH=258 |
