Probe CUST_51774_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51774_PI426222305 | JHI_St_60k_v1 | DMT400069083 | TAGTTGATTTGTACCCTGCACGTGAGGCATAATCCAAATACATTAGAGCCTACCAAAAAA |
All Microarray Probes Designed to Gene DMG400026871
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51776_PI426222305 | JHI_St_60k_v1 | DMT400069082 | GCTTGTGCCAGAAAGAATAACTTATACTTGATCGCGACTCCTCATCAACCTTGTCAAGAG |
| CUST_51774_PI426222305 | JHI_St_60k_v1 | DMT400069083 | TAGTTGATTTGTACCCTGCACGTGAGGCATAATCCAAATACATTAGAGCCTACCAAAAAA |
Microarray Signals from CUST_51774_PI426222305
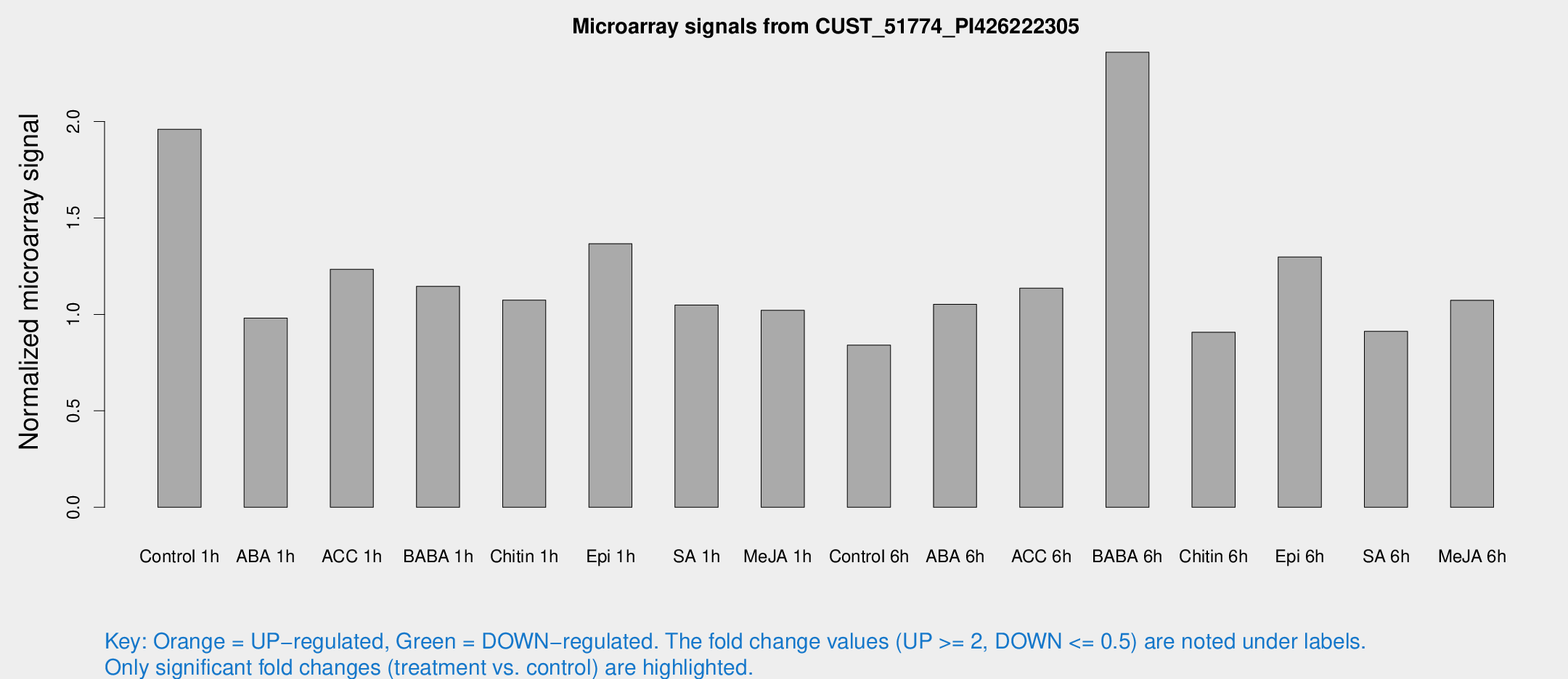
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 14.4813 | 3.49199 | 1.95984 | 0.513235 |
| ABA 1h | 6.29085 | 3.27036 | 0.980344 | 0.519 |
| ACC 1h | 9.14253 | 4.36884 | 1.23403 | 0.567186 |
| BABA 1h | 8.35626 | 3.64581 | 1.14566 | 0.558004 |
| Chitin 1h | 7.02225 | 3.38876 | 1.07408 | 0.523332 |
| Epi 1h | 9.75982 | 3.80646 | 1.36605 | 0.630494 |
| SA 1h | 8.24916 | 3.47186 | 1.04864 | 0.492496 |
| Me-JA 1h | 6.02838 | 3.51563 | 1.02081 | 0.591809 |
| Control 6h | 6.06742 | 3.52211 | 0.840414 | 0.487393 |
| ABA 6h | 8.59453 | 3.6464 | 1.05214 | 0.510205 |
| ACC 6h | 10.2318 | 4.1747 | 1.13598 | 0.538258 |
| BABA 6h | 22.5453 | 9.64901 | 2.35893 | 1.20024 |
| Chitin 6h | 6.94983 | 3.897 | 0.907054 | 0.508938 |
| Epi 6h | 11.9719 | 4.42771 | 1.29762 | 0.613624 |
| SA 6h | 6.49578 | 3.76655 | 0.912426 | 0.52891 |
| Me-JA 6h | 8.17852 | 3.53478 | 1.07312 | 0.523823 |
Source Transcript PGSC0003DMT400069083 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G26310.1 | +3 | 7e-99 | 270 | 141/331 (43%) | cytochrome P450, family 71, subfamily B, polypeptide 35 | chr3:9641089-9642779 REVERSE LENGTH=500 |
