Probe CUST_51703_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51703_PI426222305 | JHI_St_60k_v1 | DMT400066071 | TCGTTCATAAATTTCAGGGGCGAGAAAGCATTTTGTAGCACAGAGTGTCGGTACAGGCAA |
All Microarray Probes Designed to Gene DMG400025721
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51703_PI426222305 | JHI_St_60k_v1 | DMT400066071 | TCGTTCATAAATTTCAGGGGCGAGAAAGCATTTTGTAGCACAGAGTGTCGGTACAGGCAA |
| CUST_51704_PI426222305 | JHI_St_60k_v1 | DMT400066070 | GACATATATATGTATAGGGGCGAGAAAGCATTTTGTAGCACAGAGTGTCGGTACAGGCAA |
Microarray Signals from CUST_51703_PI426222305
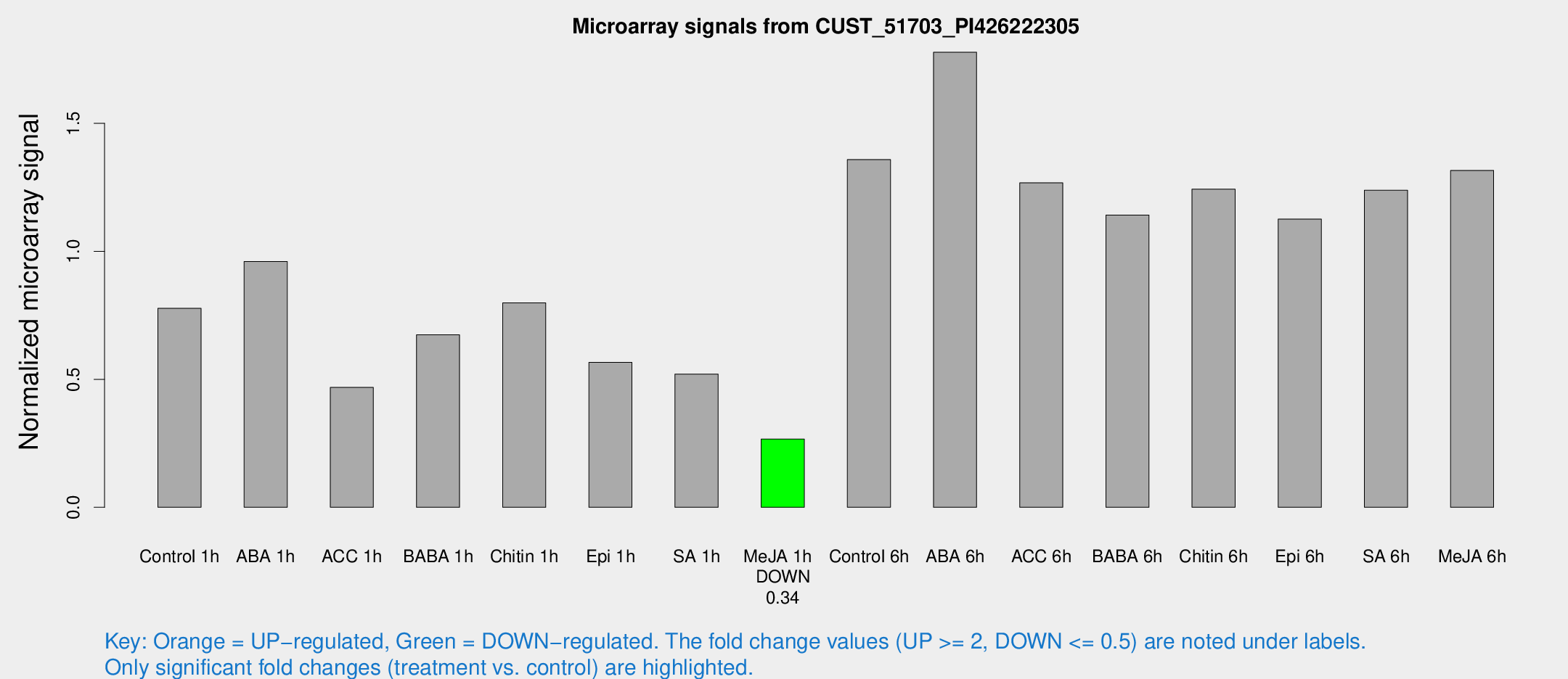
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4008.66 | 232.081 | 0.777461 | 0.0448918 |
| ABA 1h | 4445.25 | 553.824 | 0.960453 | 0.0807964 |
| ACC 1h | 2628.37 | 585.499 | 0.468234 | 0.0877618 |
| BABA 1h | 3482.45 | 662.835 | 0.673791 | 0.073954 |
| Chitin 1h | 3912.56 | 988.26 | 0.798536 | 0.136033 |
| Epi 1h | 2546.66 | 255.024 | 0.566054 | 0.0545723 |
| SA 1h | 2965.73 | 715.087 | 0.520614 | 0.12925 |
| Me-JA 1h | 1145.29 | 159.308 | 0.266899 | 0.0245446 |
| Control 6h | 7586.01 | 2134.66 | 1.35866 | 0.311692 |
| ABA 6h | 9831.83 | 1007.71 | 1.77796 | 0.112334 |
| ACC 6h | 7635.07 | 1040.17 | 1.26766 | 0.179824 |
| BABA 6h | 6743.57 | 1089.29 | 1.1419 | 0.161673 |
| Chitin 6h | 6834.83 | 445.218 | 1.24307 | 0.101116 |
| Epi 6h | 6660.51 | 889.036 | 1.12598 | 0.0957248 |
| SA 6h | 6537.73 | 1158.25 | 1.23855 | 0.0988264 |
| Me-JA 6h | 6927.59 | 1193.95 | 1.31615 | 0.135522 |
Source Transcript PGSC0003DMT400066071 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G74940.1 | +3 | 6e-29 | 94 | 73/189 (39%) | Protein of unknown function (DUF581) | chr1:28146284-28147065 FORWARD LENGTH=222 |
