Probe CUST_51561_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51561_PI426222305 | JHI_St_60k_v1 | DMT400028324 | CAAGGGATAGAGATCTTTAATTAGTTCTTGTTTGTTTGGGGACATTTCCATTTACTCGTT |
All Microarray Probes Designed to Gene DMG402010918
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51561_PI426222305 | JHI_St_60k_v1 | DMT400028324 | CAAGGGATAGAGATCTTTAATTAGTTCTTGTTTGTTTGGGGACATTTCCATTTACTCGTT |
Microarray Signals from CUST_51561_PI426222305
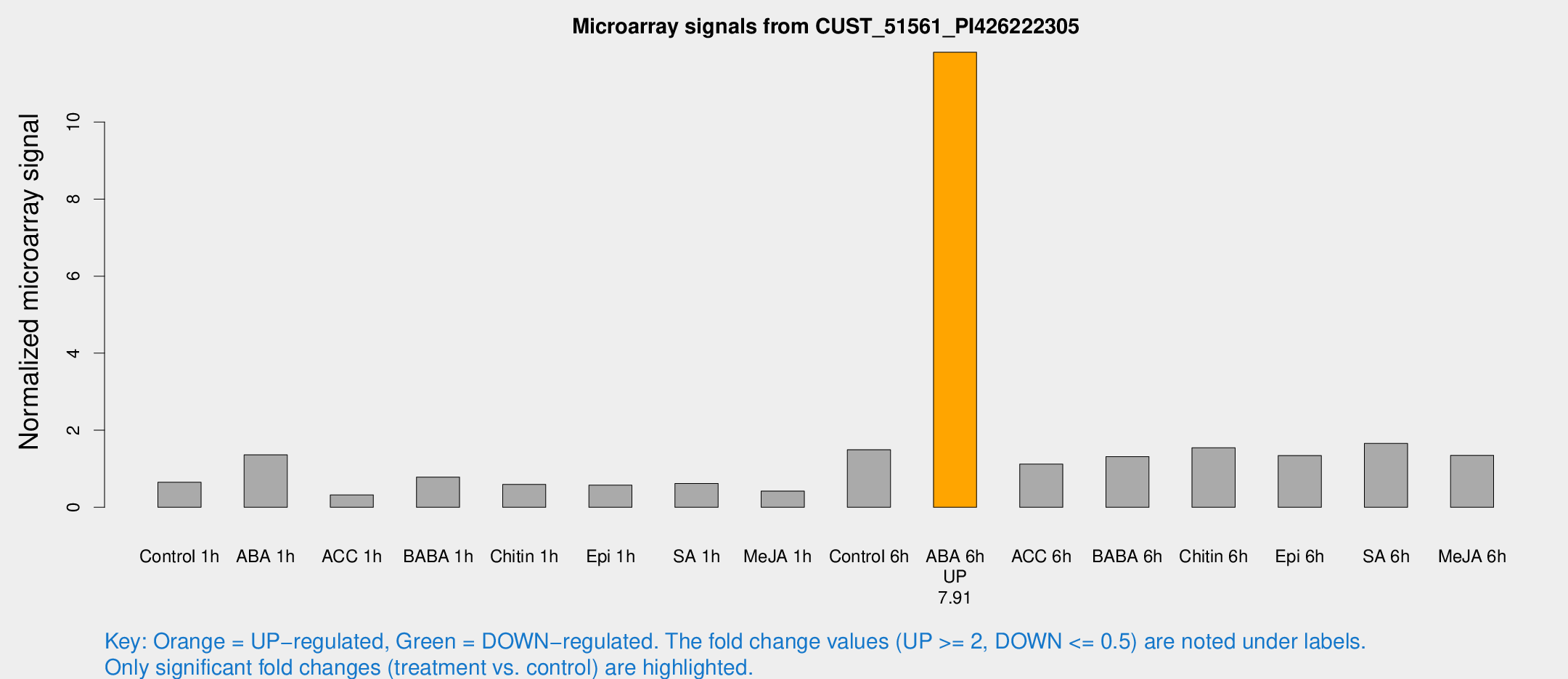
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 145.958 | 9.18807 | 0.648744 | 0.0618967 |
| ABA 1h | 314.679 | 101.721 | 1.36135 | 0.544381 |
| ACC 1h | 99.3303 | 41.0611 | 0.322431 | 0.213601 |
| BABA 1h | 176.04 | 31.1455 | 0.783591 | 0.0735421 |
| Chitin 1h | 124.509 | 24.8813 | 0.593686 | 0.087687 |
| Epi 1h | 116.402 | 22.0439 | 0.573577 | 0.117551 |
| SA 1h | 143.347 | 9.05458 | 0.620257 | 0.0596909 |
| Me-JA 1h | 82.1075 | 21.0856 | 0.421647 | 0.0755037 |
| Control 6h | 351.113 | 66.6014 | 1.4938 | 0.186316 |
| ABA 6h | 2824.15 | 163.3 | 11.8109 | 0.6821 |
| ACC 6h | 291.174 | 24.5257 | 1.12159 | 0.177608 |
| BABA 6h | 335.613 | 38.1879 | 1.31613 | 0.106614 |
| Chitin 6h | 373.486 | 36.4622 | 1.54713 | 0.183203 |
| Epi 6h | 340.883 | 20.248 | 1.34189 | 0.0985988 |
| SA 6h | 369.14 | 21.7112 | 1.65979 | 0.15044 |
| Me-JA 6h | 338.642 | 117.833 | 1.34782 | 0.362256 |
Source Transcript PGSC0003DMT400028324 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G13870.1 | +1 | 3e-154 | 438 | 199/266 (75%) | xyloglucan endotransglucosylase/hydrolase 5 | chr5:4475089-4476217 REVERSE LENGTH=293 |
