Probe CUST_51414_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51414_PI426222305 | JHI_St_60k_v1 | DMT400034336 | TTCCATCTCCTTTCTGTTTTCAGTTGGTAAAAAGGTTTGAGCAGGGACGGAGCTAGGAAG |
All Microarray Probes Designed to Gene DMG400013201
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51414_PI426222305 | JHI_St_60k_v1 | DMT400034336 | TTCCATCTCCTTTCTGTTTTCAGTTGGTAAAAAGGTTTGAGCAGGGACGGAGCTAGGAAG |
Microarray Signals from CUST_51414_PI426222305
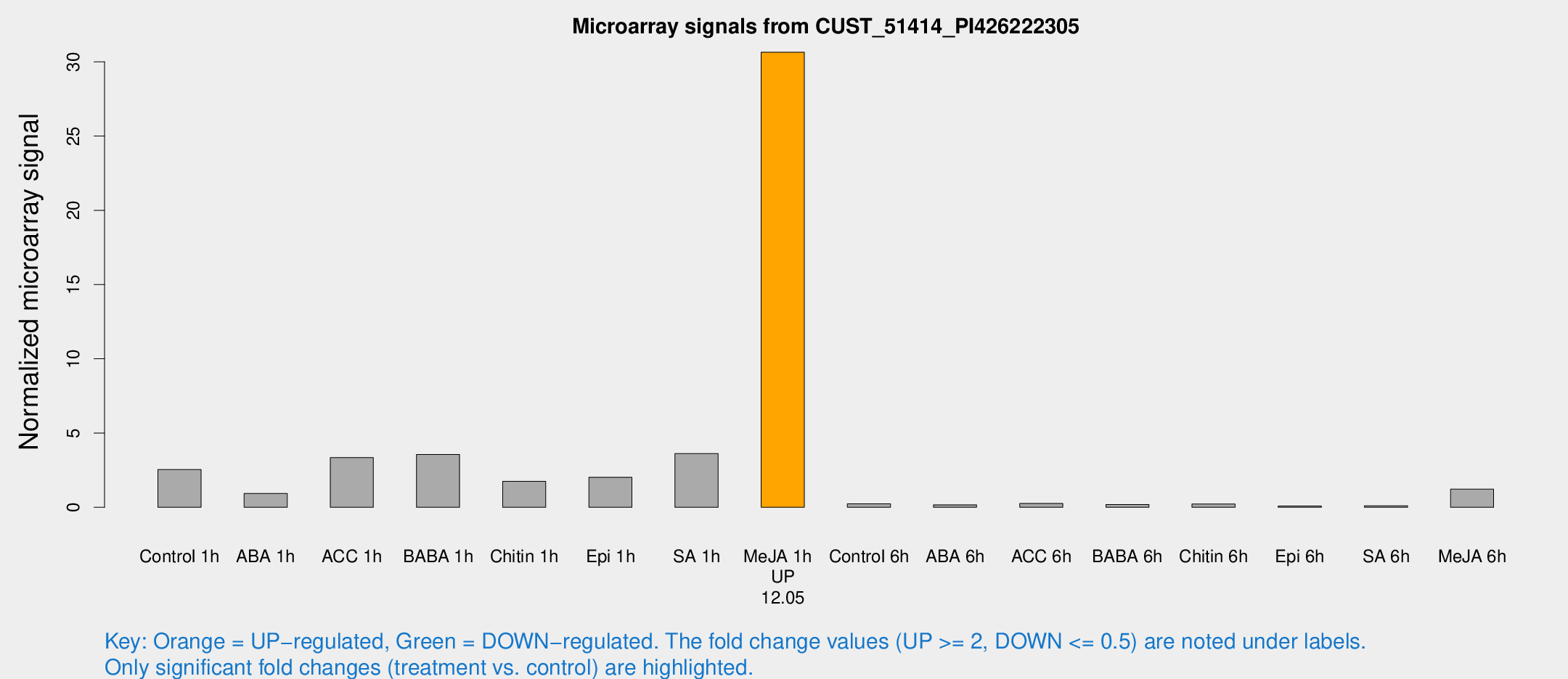
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 221.588 | 93.325 | 2.54399 | 1.09756 |
| ABA 1h | 61.2337 | 8.24236 | 0.934648 | 0.0719329 |
| ACC 1h | 329.127 | 141.622 | 3.34613 | 2.06829 |
| BABA 1h | 258.303 | 47.758 | 3.55804 | 0.739978 |
| Chitin 1h | 115.072 | 8.88631 | 1.74632 | 0.269255 |
| Epi 1h | 129.583 | 17.6152 | 2.01907 | 0.27628 |
| SA 1h | 301.417 | 102.201 | 3.61388 | 1.36833 |
| Me-JA 1h | 1849.81 | 218.187 | 30.6424 | 1.91265 |
| Control 6h | 30.2016 | 22.1601 | 0.230998 | 0.233244 |
| ABA 6h | 14.7847 | 4.83582 | 0.166251 | 0.0712002 |
| ACC 6h | 30.6449 | 15.4581 | 0.258126 | 0.245651 |
| BABA 6h | 23.9444 | 14.8847 | 0.192985 | 0.161451 |
| Chitin 6h | 17.358 | 3.68237 | 0.222408 | 0.0481625 |
| Epi 6h | 7.20535 | 3.77998 | 0.0865922 | 0.0459159 |
| SA 6h | 7.87918 | 3.41824 | 0.105948 | 0.0487758 |
| Me-JA 6h | 94.9692 | 22.4652 | 1.21921 | 0.482049 |
Source Transcript PGSC0003DMT400034336 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G33260.1 | +1 | 5e-94 | 293 | 156/324 (48%) | Protein kinase superfamily protein | chr1:12064796-12066114 FORWARD LENGTH=349 |
