Probe CUST_51367_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51367_PI426222305 | JHI_St_60k_v1 | DMT400011185 | CCATACACAATTCCCCAAATGAATAATGGACTTTGTAATAAGGGAATATCAGACCAATAG |
All Microarray Probes Designed to Gene DMG400004378
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51367_PI426222305 | JHI_St_60k_v1 | DMT400011185 | CCATACACAATTCCCCAAATGAATAATGGACTTTGTAATAAGGGAATATCAGACCAATAG |
Microarray Signals from CUST_51367_PI426222305
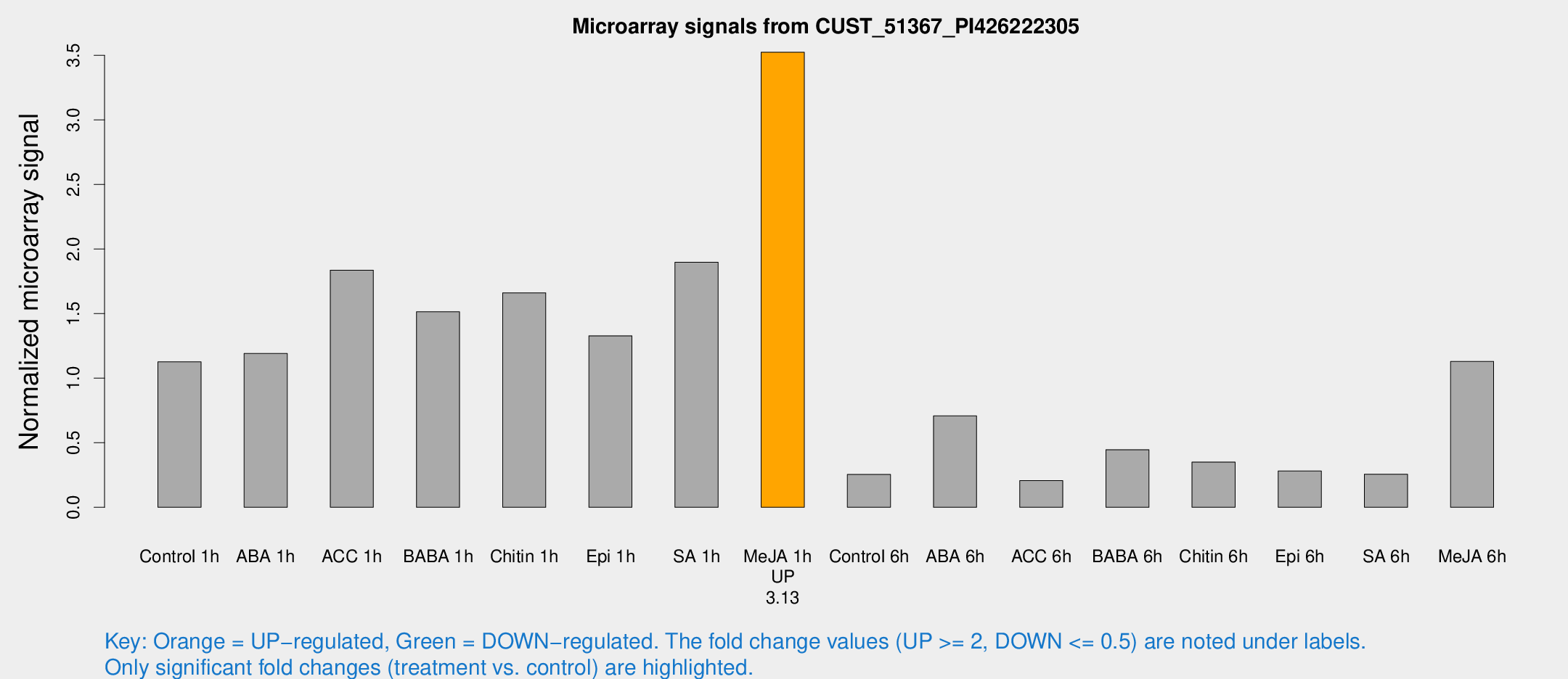
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 207.713 | 33.15 | 1.12636 | 0.137314 |
| ABA 1h | 212.479 | 76.0261 | 1.19099 | 0.361999 |
| ACC 1h | 347.048 | 56.3549 | 1.8365 | 0.446752 |
| BABA 1h | 265.032 | 30.0564 | 1.51392 | 0.0892168 |
| Chitin 1h | 279.072 | 58.477 | 1.66098 | 0.325861 |
| Epi 1h | 206.557 | 12.3 | 1.32816 | 0.0788908 |
| SA 1h | 352.106 | 33.9386 | 1.897 | 0.225399 |
| Me-JA 1h | 515.245 | 29.894 | 3.52376 | 0.328174 |
| Control 6h | 83.0965 | 48.0534 | 0.254254 | 0.368007 |
| ABA 6h | 139.751 | 24.591 | 0.708051 | 0.141094 |
| ACC 6h | 46.3679 | 12.0306 | 0.206297 | 0.0674672 |
| BABA 6h | 90.4365 | 9.70825 | 0.446065 | 0.0308591 |
| Chitin 6h | 69.9595 | 15.2879 | 0.350331 | 0.0741694 |
| Epi 6h | 58.7362 | 11.179 | 0.281172 | 0.0592133 |
| SA 6h | 45.964 | 4.88791 | 0.256584 | 0.0403101 |
| Me-JA 6h | 213.171 | 45.557 | 1.1295 | 0.204589 |
Source Transcript PGSC0003DMT400011185 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G28590.1 | +2 | 3e-137 | 404 | 207/356 (58%) | GDSL-like Lipase/Acylhydrolase superfamily protein | chr1:10047509-10049300 REVERSE LENGTH=403 |
