Probe CUST_51361_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51361_PI426222305 | JHI_St_60k_v1 | DMT400011188 | GCACATGATCTTTAGTGCTTGTTTGGTTAATGTTGTACCAAGTCAAATTAAAAGTCCTTG |
All Microarray Probes Designed to Gene DMG400004380
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51361_PI426222305 | JHI_St_60k_v1 | DMT400011188 | GCACATGATCTTTAGTGCTTGTTTGGTTAATGTTGTACCAAGTCAAATTAAAAGTCCTTG |
Microarray Signals from CUST_51361_PI426222305
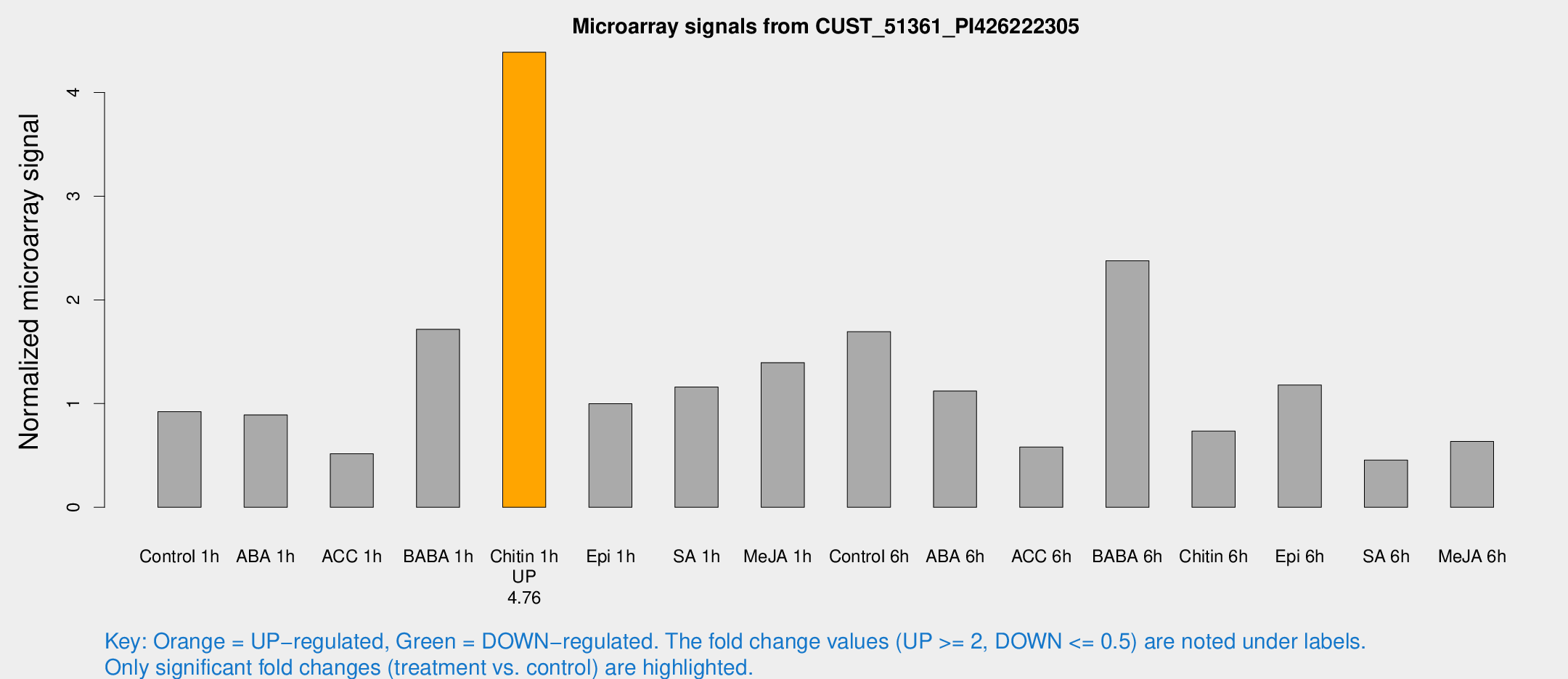
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 25.07 | 3.71975 | 0.921728 | 0.214082 |
| ABA 1h | 21.2581 | 3.51923 | 0.891118 | 0.159302 |
| ACC 1h | 14.3954 | 4.14516 | 0.515615 | 0.159241 |
| BABA 1h | 44.7833 | 5.86518 | 1.71678 | 0.179376 |
| Chitin 1h | 114.747 | 29.9839 | 4.38787 | 1.16511 |
| Epi 1h | 23.1343 | 3.55649 | 0.998312 | 0.157251 |
| SA 1h | 36.1905 | 10.9624 | 1.16052 | 0.437122 |
| Me-JA 1h | 59.4267 | 45.8463 | 1.39509 | 2.03762 |
| Control 6h | 51.6401 | 19.3679 | 1.69404 | 0.700925 |
| ABA 6h | 37.2727 | 12.1457 | 1.12103 | 0.675985 |
| ACC 6h | 24.5242 | 13.6641 | 0.581321 | 0.312747 |
| BABA 6h | 101.843 | 44.9495 | 2.37742 | 2.7734 |
| Chitin 6h | 40.7042 | 31.0402 | 0.734933 | 0.995974 |
| Epi 6h | 38.6485 | 10.567 | 1.17917 | 0.293932 |
| SA 6h | 13.0696 | 3.89892 | 0.454854 | 0.168127 |
| Me-JA 6h | 33.6772 | 26.1143 | 0.634673 | 0.770479 |
Source Transcript PGSC0003DMT400011188 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G49340.1 | +2 | 4e-113 | 338 | 164/339 (48%) | Cysteine proteinases superfamily protein | chr3:18293347-18294577 REVERSE LENGTH=341 |
