Probe CUST_51249_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51249_PI426222305 | JHI_St_60k_v1 | DMT400006026 | CTTATTAGTAGCTAGTCAACAAGAACAACCGTCAGACGTCTCAGCCCAAAATTTCCTATT |
All Microarray Probes Designed to Gene DMG400002341
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51249_PI426222305 | JHI_St_60k_v1 | DMT400006026 | CTTATTAGTAGCTAGTCAACAAGAACAACCGTCAGACGTCTCAGCCCAAAATTTCCTATT |
Microarray Signals from CUST_51249_PI426222305
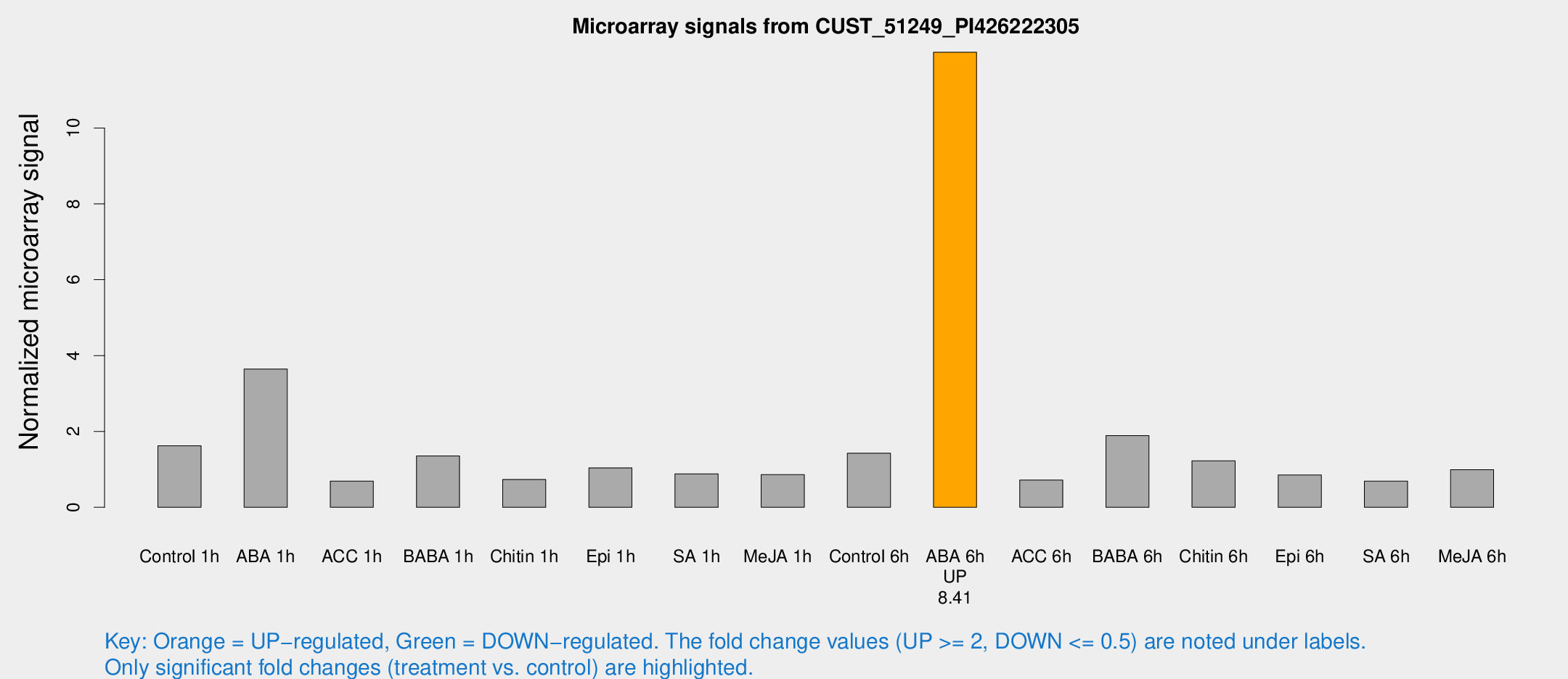
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 15.8634 | 3.34257 | 1.62207 | 0.364135 |
| ABA 1h | 40.3166 | 15.6002 | 3.64604 | 2.53989 |
| ACC 1h | 6.74765 | 3.60646 | 0.690531 | 0.370503 |
| BABA 1h | 14.7062 | 5.80054 | 1.35256 | 0.500821 |
| Chitin 1h | 6.27086 | 3.20626 | 0.733314 | 0.373977 |
| Epi 1h | 8.87807 | 3.18637 | 1.03735 | 0.418574 |
| SA 1h | 9.03498 | 3.196 | 0.882277 | 0.35515 |
| Me-JA 1h | 7.13264 | 3.2802 | 0.860723 | 0.433024 |
| Control 6h | 14.8943 | 4.40665 | 1.42713 | 0.427804 |
| ABA 6h | 125.974 | 23.6384 | 11.997 | 2.11687 |
| ACC 6h | 7.95672 | 4.02823 | 0.717088 | 0.364548 |
| BABA 6h | 48.6937 | 40.6882 | 1.8913 | 3.83313 |
| Chitin 6h | 12.9683 | 3.7518 | 1.22502 | 0.395057 |
| Epi 6h | 10.2763 | 3.84265 | 0.851357 | 0.399055 |
| SA 6h | 6.49188 | 3.4843 | 0.689502 | 0.373849 |
| Me-JA 6h | 10.0565 | 3.32022 | 0.992939 | 0.387547 |
Source Transcript PGSC0003DMT400006026 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G26300.1 | +3 | 1e-77 | 226 | 121/292 (41%) | cytochrome P450, family 71, subfamily B, polypeptide 34 | chr3:9639199-9640866 REVERSE LENGTH=500 |
