Probe CUST_51239_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51239_PI426222305 | JHI_St_60k_v1 | DMT400085105 | TATTTTGATGGACTCATGCCTCACAAAAGAAGAAGGCCAAAGAAGAGAGTTCAGAGATGA |
All Microarray Probes Designed to Gene DMG400034676
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51239_PI426222305 | JHI_St_60k_v1 | DMT400085105 | TATTTTGATGGACTCATGCCTCACAAAAGAAGAAGGCCAAAGAAGAGAGTTCAGAGATGA |
Microarray Signals from CUST_51239_PI426222305
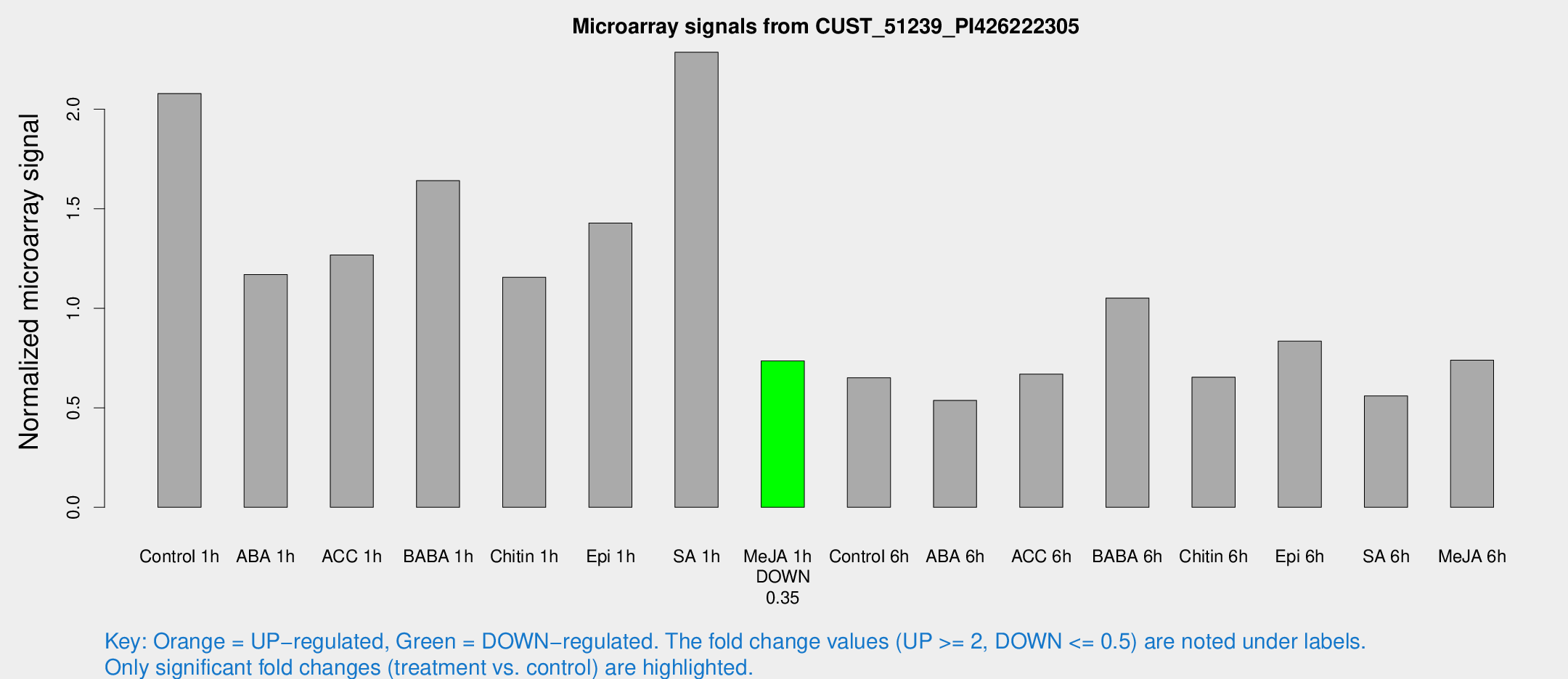
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8063.45 | 1820.07 | 2.07878 | 0.323155 |
| ABA 1h | 3845.45 | 229.092 | 1.16963 | 0.0675367 |
| ACC 1h | 5947.39 | 2121.74 | 1.26732 | 0.636562 |
| BABA 1h | 6294.27 | 1567.59 | 1.64102 | 0.300642 |
| Chitin 1h | 3865.77 | 256.624 | 1.15593 | 0.0667458 |
| Epi 1h | 4586.97 | 265.377 | 1.42824 | 0.082467 |
| SA 1h | 8901.29 | 1378.28 | 2.28616 | 0.208254 |
| Me-JA 1h | 2259.81 | 299.097 | 0.735191 | 0.0424633 |
| Control 6h | 2515.89 | 463.14 | 0.651141 | 0.0764181 |
| ABA 6h | 2245.22 | 521.307 | 0.537122 | 0.108993 |
| ACC 6h | 2849.85 | 164.951 | 0.669364 | 0.080783 |
| BABA 6h | 4388.46 | 376.072 | 1.05114 | 0.0606963 |
| Chitin 6h | 2613.97 | 306.766 | 0.653862 | 0.0706694 |
| Epi 6h | 3552.43 | 465.503 | 0.834683 | 0.0698695 |
| SA 6h | 2507.6 | 1064.48 | 0.559679 | 0.222188 |
| Me-JA 6h | 3061.27 | 1032.26 | 0.738993 | 0.20119 |
Source Transcript PGSC0003DMT400085105 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G33050.1 | +1 | 5e-34 | 130 | 85/198 (43%) | receptor like protein 26 | chr2:14021870-14024272 FORWARD LENGTH=800 |
