Probe CUST_51133_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51133_PI426222305 | JHI_St_60k_v1 | DMT400019836 | TAATTTCGTGAAGAAGAAGAAGAAGCCAAAGAATTCGCCGGAGAAGAAGAGATTACCGGC |
All Microarray Probes Designed to Gene DMG400007670
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51130_PI426222305 | JHI_St_60k_v1 | DMT400019835 | TAATTTCGTGAAGAAGAAGAAGAAGCCAAAGAATTCGCCGGAGAAGAAGAGATTACCGGC |
| CUST_51133_PI426222305 | JHI_St_60k_v1 | DMT400019836 | TAATTTCGTGAAGAAGAAGAAGAAGCCAAAGAATTCGCCGGAGAAGAAGAGATTACCGGC |
Microarray Signals from CUST_51133_PI426222305
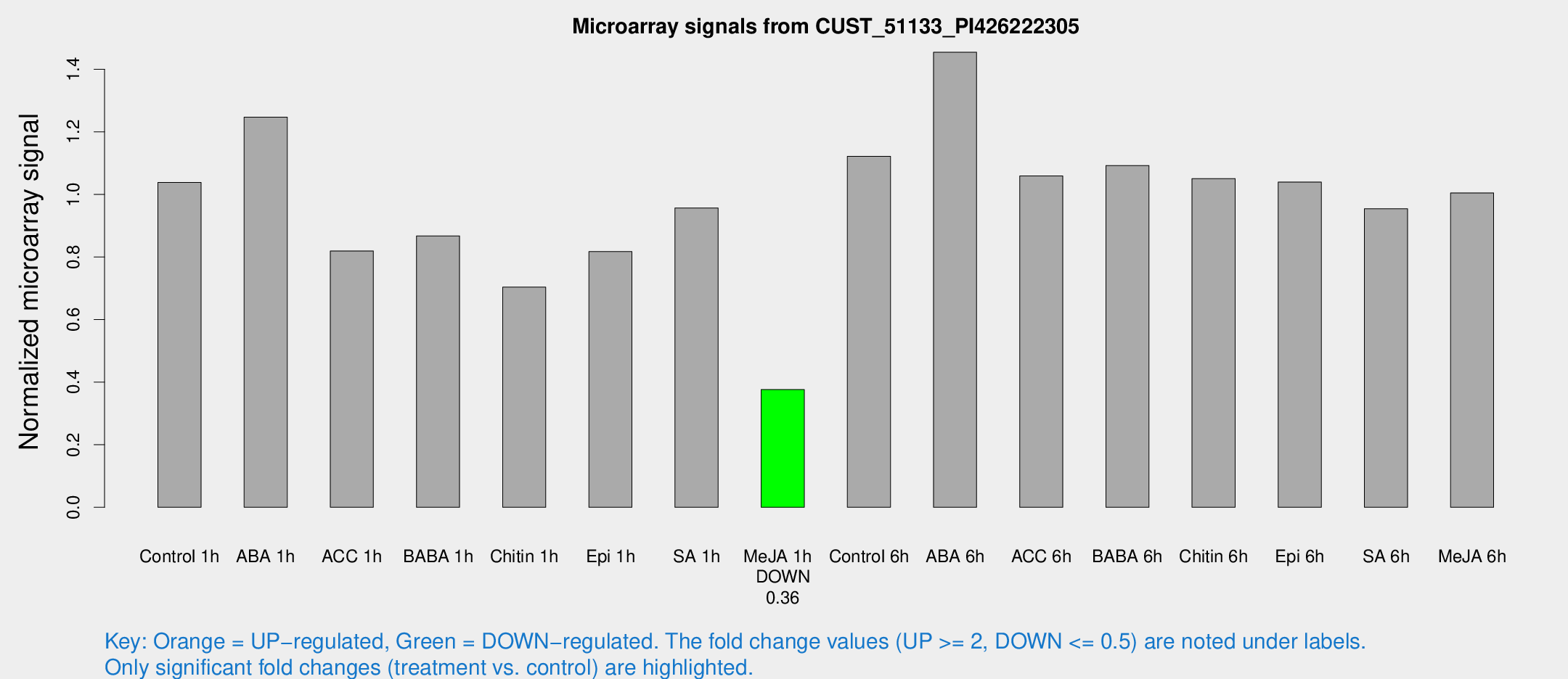
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1488.55 | 185.287 | 1.03833 | 0.082422 |
| ABA 1h | 1624.72 | 313.759 | 1.24653 | 0.184371 |
| ACC 1h | 1316.36 | 359.353 | 0.819119 | 0.228387 |
| BABA 1h | 1283.82 | 326.436 | 0.867271 | 0.179434 |
| Chitin 1h | 952.583 | 241.462 | 0.703688 | 0.156353 |
| Epi 1h | 1038.81 | 187.891 | 0.817195 | 0.159402 |
| SA 1h | 1411.02 | 182.996 | 0.956674 | 0.0644394 |
| Me-JA 1h | 443.821 | 69.3578 | 0.376141 | 0.0282907 |
| Control 6h | 1686.21 | 432.511 | 1.12132 | 0.241595 |
| ABA 6h | 2238.84 | 330.246 | 1.4542 | 0.180217 |
| ACC 6h | 1740.01 | 195.834 | 1.05879 | 0.123588 |
| BABA 6h | 1749.64 | 190.887 | 1.09225 | 0.0903286 |
| Chitin 6h | 1584.69 | 91.8475 | 1.05053 | 0.0606989 |
| Epi 6h | 1760.42 | 391.448 | 1.0393 | 0.20137 |
| SA 6h | 1632.24 | 672.565 | 0.953921 | 0.384005 |
| Me-JA 6h | 1481.51 | 312.152 | 1.00461 | 0.137132 |
Source Transcript PGSC0003DMT400019836 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G17900.1 | +1 | 1e-95 | 290 | 152/240 (63%) | PLATZ transcription factor family protein | chr4:9946046-9947697 FORWARD LENGTH=227 |
