Probe CUST_51130_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51130_PI426222305 | JHI_St_60k_v1 | DMT400019835 | TAATTTCGTGAAGAAGAAGAAGAAGCCAAAGAATTCGCCGGAGAAGAAGAGATTACCGGC |
All Microarray Probes Designed to Gene DMG400007670
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51133_PI426222305 | JHI_St_60k_v1 | DMT400019836 | TAATTTCGTGAAGAAGAAGAAGAAGCCAAAGAATTCGCCGGAGAAGAAGAGATTACCGGC |
| CUST_51130_PI426222305 | JHI_St_60k_v1 | DMT400019835 | TAATTTCGTGAAGAAGAAGAAGAAGCCAAAGAATTCGCCGGAGAAGAAGAGATTACCGGC |
Microarray Signals from CUST_51130_PI426222305
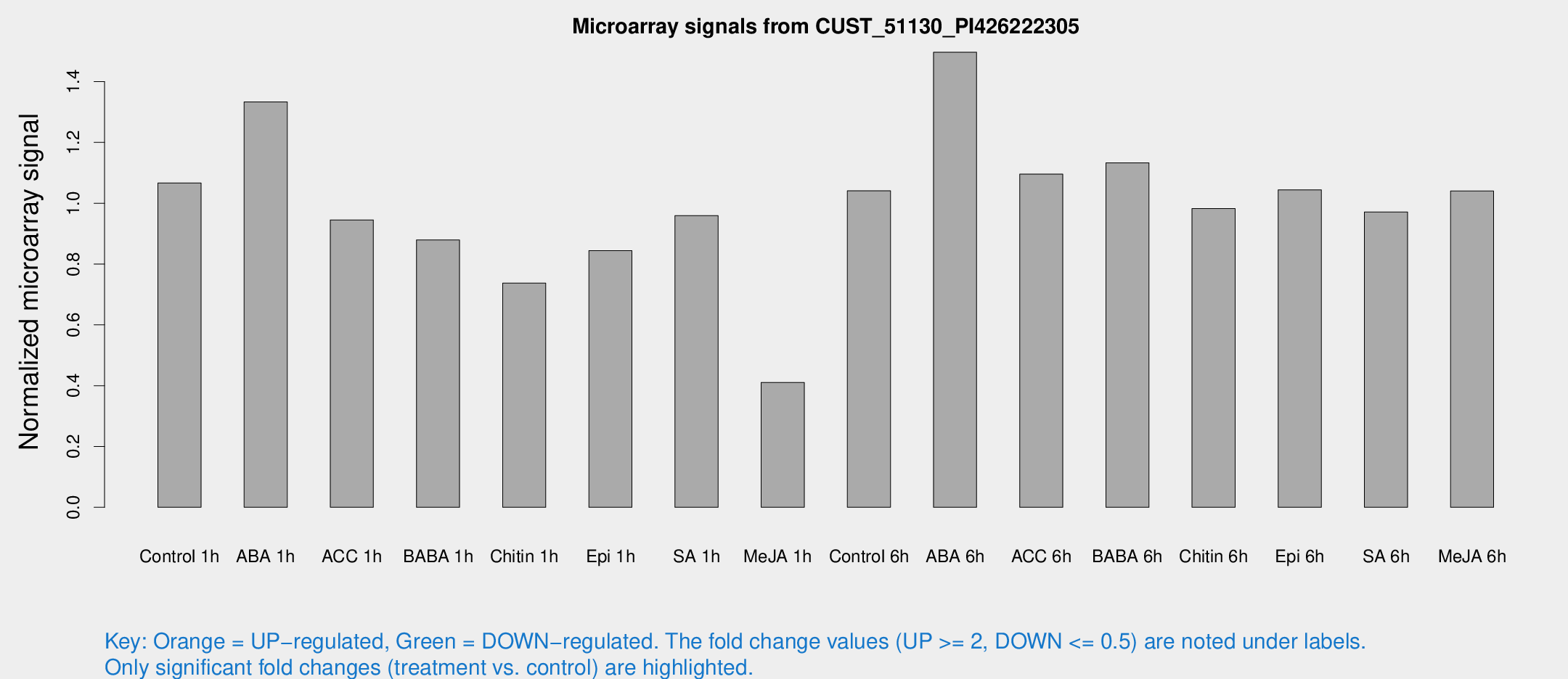
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1444.83 | 135.455 | 1.06669 | 0.061648 |
| ABA 1h | 1653.45 | 324.978 | 1.33339 | 0.196369 |
| ACC 1h | 1414.9 | 352.863 | 0.945228 | 0.220704 |
| BABA 1h | 1240.15 | 319.809 | 0.879299 | 0.185891 |
| Chitin 1h | 928.141 | 192.163 | 0.737353 | 0.122177 |
| Epi 1h | 1012.31 | 165.367 | 0.844198 | 0.143638 |
| SA 1h | 1340.91 | 157.114 | 0.959319 | 0.0600382 |
| Me-JA 1h | 464.767 | 85.802 | 0.410653 | 0.0403579 |
| Control 6h | 1510.7 | 428.717 | 1.04078 | 0.243539 |
| ABA 6h | 2204.31 | 359.639 | 1.49665 | 0.211438 |
| ACC 6h | 1714.85 | 205.931 | 1.09558 | 0.102081 |
| BABA 6h | 1724.7 | 185.13 | 1.13302 | 0.0920438 |
| Chitin 6h | 1410.2 | 90.4908 | 0.982368 | 0.056798 |
| Epi 6h | 1646.94 | 305.737 | 1.04399 | 0.146892 |
| SA 6h | 1576.33 | 640.356 | 0.971198 | 0.388511 |
| Me-JA 6h | 1464.84 | 317.145 | 1.04027 | 0.154743 |
Source Transcript PGSC0003DMT400019835 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G17900.1 | +2 | 2e-67 | 137 | 65/75 (87%) | PLATZ transcription factor family protein | chr4:9946046-9947697 FORWARD LENGTH=227 |
