Probe CUST_50910_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50910_PI426222305 | JHI_St_60k_v1 | DMT400059675 | ATTGCCATAACATATGTGAGTAGTGAAAGGAGTTGAAACTCTCTTTTGGTTGTCATATGT |
All Microarray Probes Designed to Gene DMG400023197
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50910_PI426222305 | JHI_St_60k_v1 | DMT400059675 | ATTGCCATAACATATGTGAGTAGTGAAAGGAGTTGAAACTCTCTTTTGGTTGTCATATGT |
| CUST_50906_PI426222305 | JHI_St_60k_v1 | DMT400059674 | CTTTTGTAGGTATTGCAGATGCACTAGGTGCTGCCAATGGTCTTTCTCTTGACCTCTTTG |
Microarray Signals from CUST_50910_PI426222305
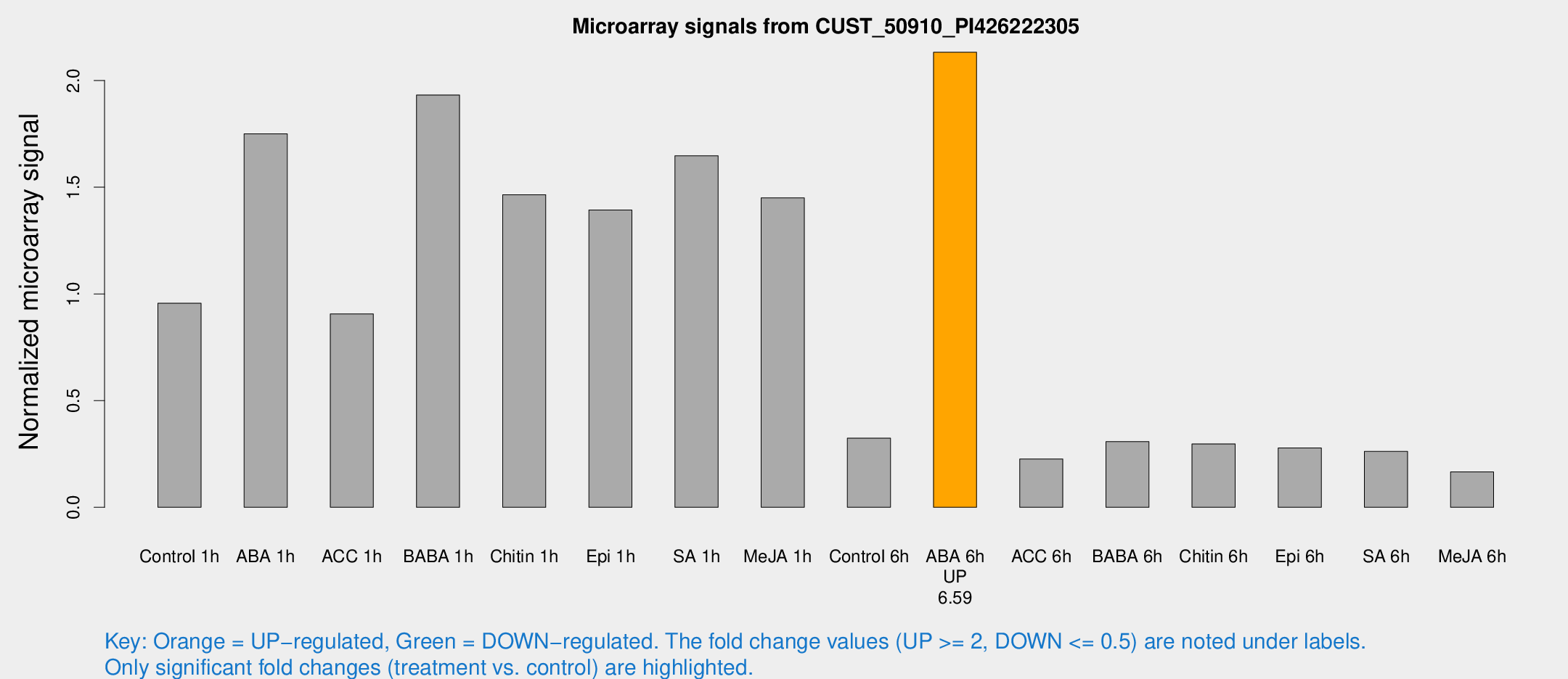
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 254.48 | 15.1352 | 0.9562 | 0.0567596 |
| ABA 1h | 429.545 | 83.0446 | 1.75006 | 0.285339 |
| ACC 1h | 285.494 | 91.9584 | 0.906238 | 0.327023 |
| BABA 1h | 541.923 | 157.543 | 1.93183 | 0.437532 |
| Chitin 1h | 360.776 | 66.9464 | 1.46468 | 0.161543 |
| Epi 1h | 320.82 | 18.8928 | 1.39307 | 0.0952244 |
| SA 1h | 452.452 | 40.1012 | 1.64704 | 0.0959854 |
| Me-JA 1h | 322.613 | 54.9918 | 1.44958 | 0.142301 |
| Control 6h | 93.9482 | 24.8024 | 0.323622 | 0.0753378 |
| ABA 6h | 619.946 | 103.886 | 2.13233 | 0.262395 |
| ACC 6h | 70.2972 | 9.11569 | 0.226656 | 0.0226365 |
| BABA 6h | 99.6689 | 26.3317 | 0.307491 | 0.0903296 |
| Chitin 6h | 85.8025 | 12.6434 | 0.297193 | 0.050217 |
| Epi 6h | 84.2299 | 9.49051 | 0.277958 | 0.053912 |
| SA 6h | 80.4439 | 33.2113 | 0.261888 | 0.0899957 |
| Me-JA 6h | 52.0864 | 22.4266 | 0.165718 | 0.0592305 |
Source Transcript PGSC0003DMT400059675 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
