Probe CUST_50655_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50655_PI426222305 | JHI_St_60k_v1 | DMT400024931 | GTTGCTCATCTTCTCAGAAACAATGATGAACCAACACCAAGATGAAGCCCGTCTATTCAG |
All Microarray Probes Designed to Gene DMG400009634
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50655_PI426222305 | JHI_St_60k_v1 | DMT400024931 | GTTGCTCATCTTCTCAGAAACAATGATGAACCAACACCAAGATGAAGCCCGTCTATTCAG |
| CUST_50643_PI426222305 | JHI_St_60k_v1 | DMT400024930 | ATCTCACAAGAACTCTTGAAAGCTATCGAAGAGTCTCAAGTTGCACTCATCGTTTTCTGA |
Microarray Signals from CUST_50655_PI426222305
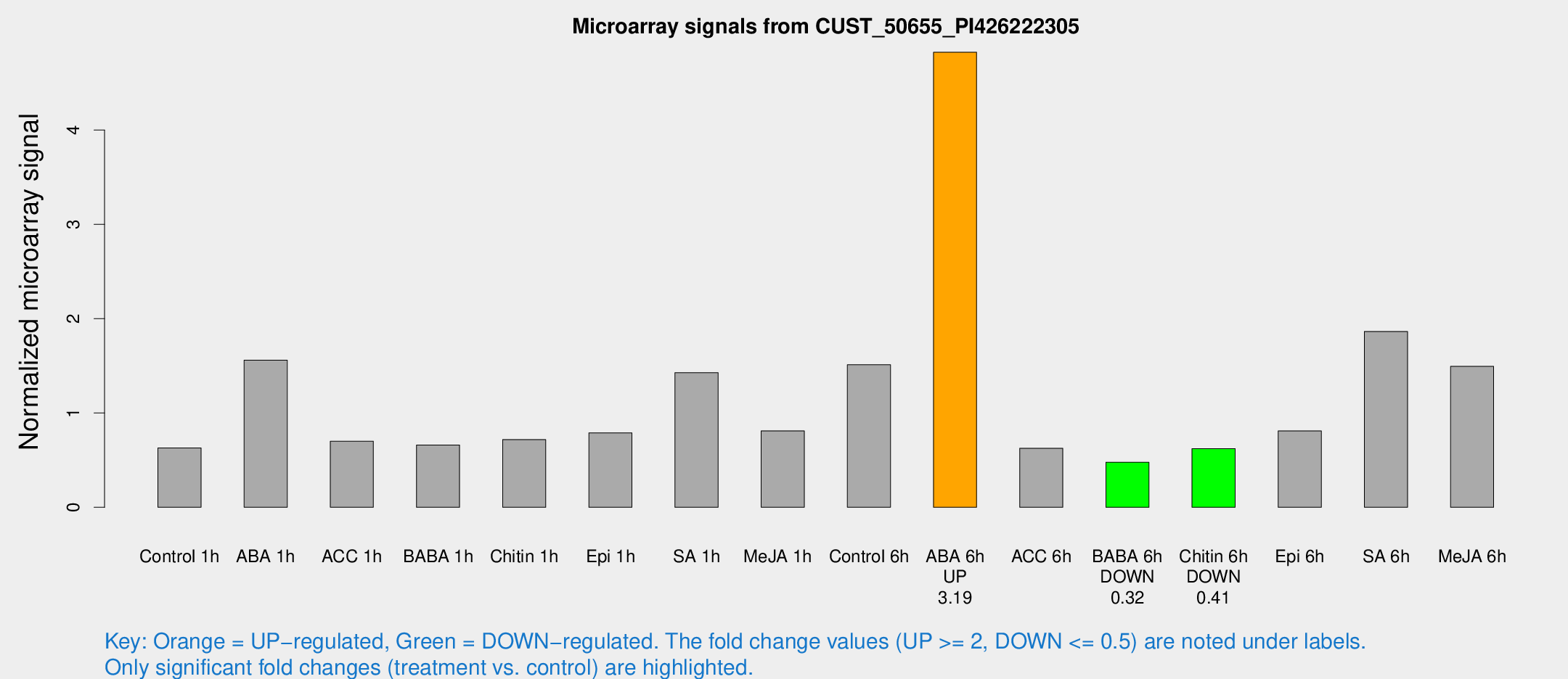
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.72144 | 3.34537 | 0.62979 | 0.284451 |
| ABA 1h | 19.5049 | 6.11101 | 1.56017 | 0.48708 |
| ACC 1h | 10.1066 | 4.13494 | 0.700243 | 0.331833 |
| BABA 1h | 9.00517 | 3.56862 | 0.660302 | 0.313669 |
| Chitin 1h | 8.90521 | 3.33658 | 0.718131 | 0.317661 |
| Epi 1h | 9.39762 | 3.24903 | 0.789753 | 0.312589 |
| SA 1h | 19.5789 | 3.69432 | 1.42708 | 0.279013 |
| Me-JA 1h | 8.96546 | 3.42497 | 0.810339 | 0.333846 |
| Control 6h | 20.5241 | 4.19595 | 1.51189 | 0.308653 |
| ABA 6h | 67.4936 | 8.53336 | 4.82531 | 0.478095 |
| ACC 6h | 10.2421 | 4.0568 | 0.626061 | 0.294315 |
| BABA 6h | 6.99185 | 3.79409 | 0.477754 | 0.262054 |
| Chitin 6h | 8.67802 | 3.78124 | 0.622662 | 0.28146 |
| Epi 6h | 12.4838 | 4.03129 | 0.810106 | 0.299385 |
| SA 6h | 24.7799 | 4.5645 | 1.86485 | 0.600089 |
| Me-JA 6h | 22.8249 | 7.57216 | 1.49391 | 0.633211 |
Source Transcript PGSC0003DMT400024931 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G72860.1 | +1 | 6e-32 | 130 | 60/108 (56%) | Disease resistance protein (TIR-NBS-LRR class) family | chr1:27417096-27420778 REVERSE LENGTH=1163 |
