Probe CUST_50588_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50588_PI426222305 | JHI_St_60k_v1 | DMT400022364 | CATACAGTCCATCCGTTAAAAAGGACCCGTTTGACCCAAAAATACTTGGGAAGTGGTGTT |
All Microarray Probes Designed to Gene DMG400008677
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50593_PI426222305 | JHI_St_60k_v1 | DMT400022365 | ATAAGGATCATGACATAGTACTCTGCAAAGTGTATAGAAAGGCAACCTCCTTTAAAGTAT |
| CUST_50588_PI426222305 | JHI_St_60k_v1 | DMT400022364 | CATACAGTCCATCCGTTAAAAAGGACCCGTTTGACCCAAAAATACTTGGGAAGTGGTGTT |
| CUST_50586_PI426222305 | JHI_St_60k_v1 | DMT400022366 | AGGGGATTCAAGAATTCGCTTGGACGTTCATGGGTTCAATTCTGTGCAATAACACTTTCC |
Microarray Signals from CUST_50588_PI426222305
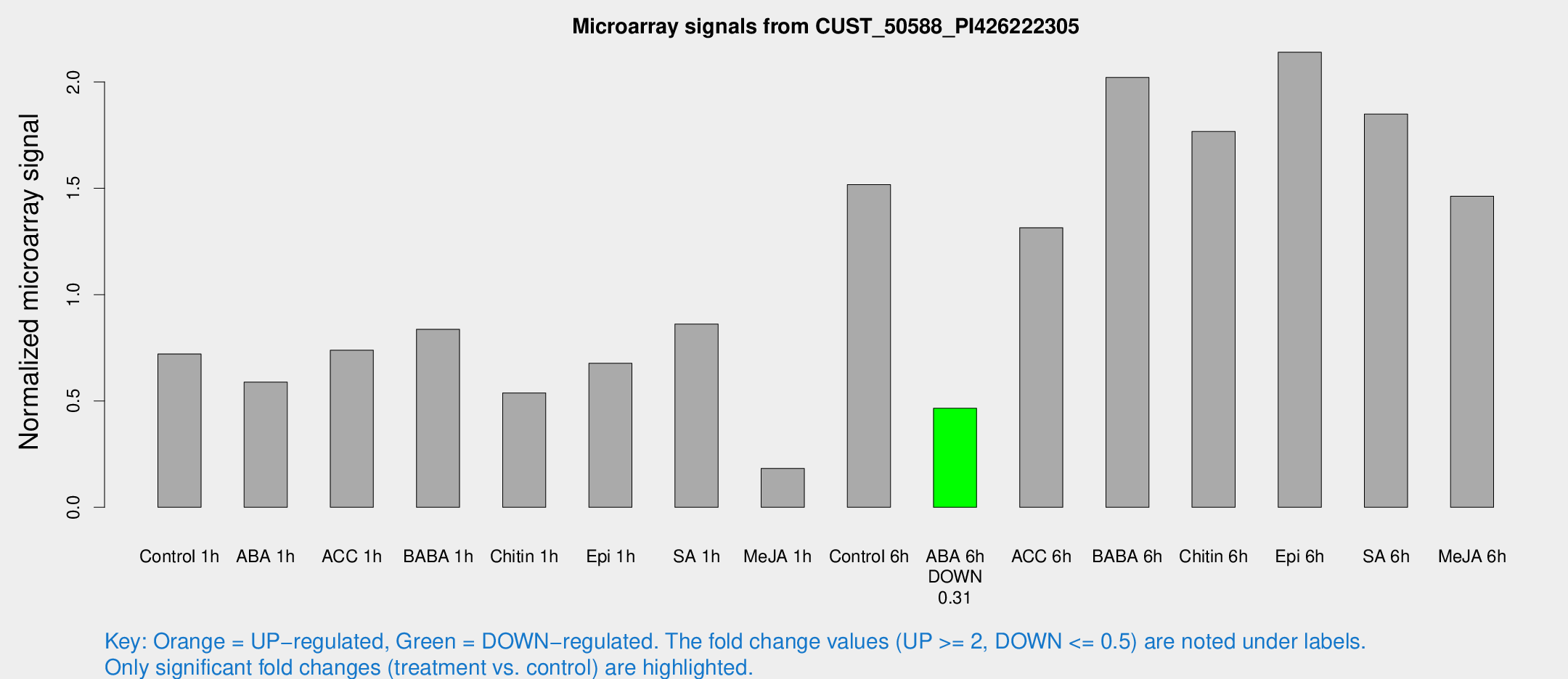
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 233.714 | 43.5491 | 0.721135 | 0.0840988 |
| ABA 1h | 171.433 | 38.2431 | 0.588998 | 0.0870664 |
| ACC 1h | 286.566 | 97.5704 | 0.738767 | 0.333093 |
| BABA 1h | 270.325 | 65.592 | 0.837117 | 0.142812 |
| Chitin 1h | 162.304 | 44.1523 | 0.537779 | 0.105723 |
| Epi 1h | 193.189 | 40.4645 | 0.676965 | 0.154647 |
| SA 1h | 296.942 | 79.2859 | 0.861162 | 0.172797 |
| Me-JA 1h | 47.5218 | 5.47843 | 0.182991 | 0.033347 |
| Control 6h | 515.31 | 127.007 | 1.51728 | 0.305367 |
| ABA 6h | 159.876 | 27.5157 | 0.465675 | 0.0526277 |
| ACC 6h | 479.097 | 51.4864 | 1.31445 | 0.186201 |
| BABA 6h | 711.561 | 41.401 | 2.0209 | 0.117336 |
| Chitin 6h | 604.6 | 95.3128 | 1.76698 | 0.315709 |
| Epi 6h | 765.167 | 80.4499 | 2.13971 | 0.247148 |
| SA 6h | 614.535 | 142.276 | 1.84868 | 0.294311 |
| Me-JA 6h | 639.178 | 369.461 | 1.46229 | 0.868141 |
Source Transcript PGSC0003DMT400022364 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
