Probe CUST_50539_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50539_PI426222305 | JHI_St_60k_v1 | DMT400062402 | GTGGGGTTTTTAATCAGAAGGCTAATCGCAAGGTTGTTAATTAGGAAACATATGTTATAC |
All Microarray Probes Designed to Gene DMG400024285
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50539_PI426222305 | JHI_St_60k_v1 | DMT400062402 | GTGGGGTTTTTAATCAGAAGGCTAATCGCAAGGTTGTTAATTAGGAAACATATGTTATAC |
| CUST_50525_PI426222305 | JHI_St_60k_v1 | DMT400062403 | GTGGGGTTTTTAATCAGAAGGCTAATCGCAAGGTTGTTAATTAGGAAACATATGTTATAC |
Microarray Signals from CUST_50539_PI426222305
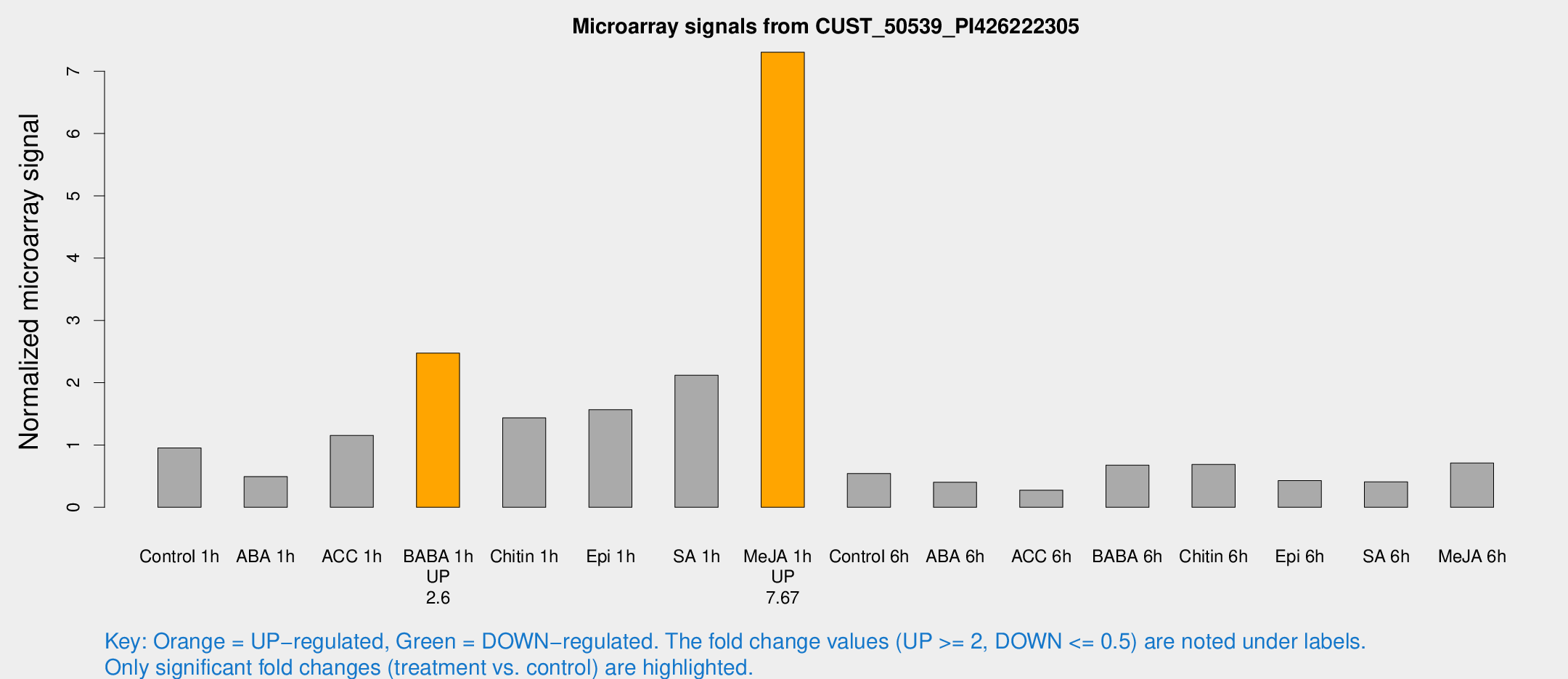
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 26.5984 | 4.70887 | 0.952756 | 0.245731 |
| ABA 1h | 12.8243 | 3.6206 | 0.492207 | 0.173531 |
| ACC 1h | 35.4763 | 10.1503 | 1.15374 | 0.329347 |
| BABA 1h | 64.8497 | 5.9565 | 2.47387 | 0.443278 |
| Chitin 1h | 35.4335 | 5.035 | 1.43492 | 0.168424 |
| Epi 1h | 36.9308 | 4.1174 | 1.56681 | 0.178872 |
| SA 1h | 83.2415 | 48.7844 | 2.12123 | 1.55789 |
| Me-JA 1h | 161.94 | 13.7202 | 7.30458 | 0.823426 |
| Control 6h | 17.1738 | 6.54021 | 0.542374 | 0.189579 |
| ABA 6h | 14.604 | 7.37441 | 0.402302 | 0.25536 |
| ACC 6h | 8.62347 | 4.48111 | 0.275749 | 0.142552 |
| BABA 6h | 26.0542 | 10.2496 | 0.676601 | 0.424951 |
| Chitin 6h | 23.6384 | 9.18761 | 0.688078 | 0.379547 |
| Epi 6h | 13.239 | 4.86364 | 0.429557 | 0.166602 |
| SA 6h | 12.6936 | 5.24825 | 0.407841 | 0.248448 |
| Me-JA 6h | 22.4288 | 7.56963 | 0.710126 | 0.27864 |
Source Transcript PGSC0003DMT400062402 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G26010.1 | +2 | 3e-111 | 332 | 171/313 (55%) | Peroxidase superfamily protein | chr4:13200653-13201688 FORWARD LENGTH=310 |
