Probe CUST_50510_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50510_PI426222305 | JHI_St_60k_v1 | DMT400062084 | GGACTCGCTTGGCCATGAGAATTATTCACTTTTTGGAATAATTTCTCGTTTTACAGAAAT |
All Microarray Probes Designed to Gene DMG400024160
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50510_PI426222305 | JHI_St_60k_v1 | DMT400062084 | GGACTCGCTTGGCCATGAGAATTATTCACTTTTTGGAATAATTTCTCGTTTTACAGAAAT |
Microarray Signals from CUST_50510_PI426222305
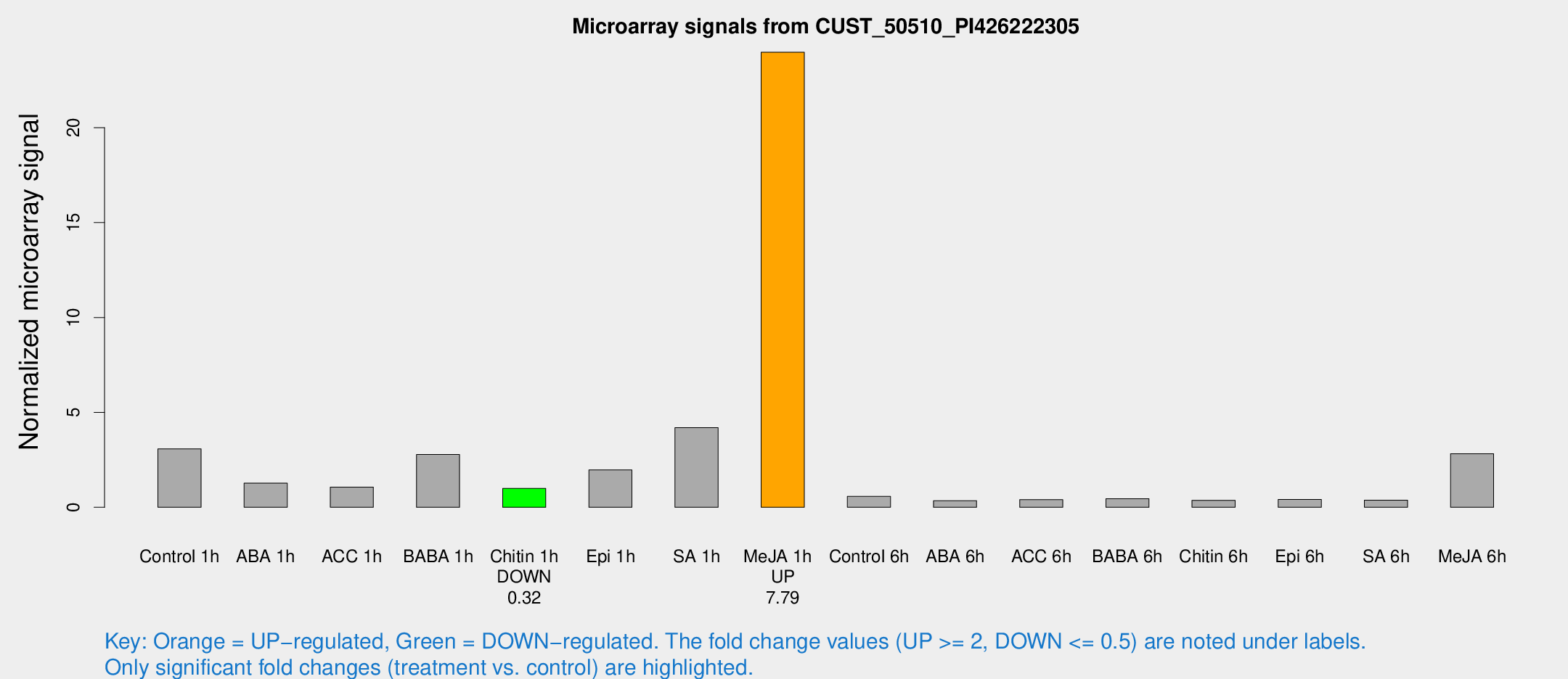
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 61.7164 | 12.5 | 3.07701 | 0.404447 |
| ABA 1h | 26.0003 | 11.4872 | 1.27183 | 0.481683 |
| ACC 1h | 24.6873 | 9.1051 | 1.06441 | 0.404812 |
| BABA 1h | 52.9545 | 8.43752 | 2.78171 | 0.385933 |
| Chitin 1h | 17.6989 | 3.76174 | 0.997459 | 0.238797 |
| Epi 1h | 33.7831 | 5.75997 | 1.9696 | 0.340107 |
| SA 1h | 94.7321 | 34.1775 | 4.19492 | 1.83504 |
| Me-JA 1h | 384.762 | 54.6404 | 23.9763 | 1.76426 |
| Control 6h | 15.1158 | 8.5536 | 0.576154 | 0.336362 |
| ABA 6h | 7.10674 | 4.12174 | 0.346188 | 0.200584 |
| ACC 6h | 9.25051 | 4.53812 | 0.404216 | 0.204076 |
| BABA 6h | 10.9238 | 4.43225 | 0.456426 | 0.214818 |
| Chitin 6h | 7.63885 | 4.4445 | 0.370566 | 0.214813 |
| Epi 6h | 8.98924 | 4.67086 | 0.410911 | 0.20667 |
| SA 6h | 7.13945 | 4.14132 | 0.373837 | 0.21668 |
| Me-JA 6h | 56.2172 | 10.8869 | 2.82563 | 0.767062 |
Source Transcript PGSC0003DMT400062084 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G16820.1 | +1 | 0.0 | 557 | 309/509 (61%) | alpha/beta-Hydrolases superfamily protein | chr4:9467563-9469116 FORWARD LENGTH=517 |
