Probe CUST_5050_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5050_PI426222305 | JHI_St_60k_v1 | DMT400009189 | CGTCATTCAAAGCCTGTTGTATATTTTTGTGAGGCATAGTGGGAAAGAGATTTTTAAAAC |
All Microarray Probes Designed to Gene DMG400003565
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5050_PI426222305 | JHI_St_60k_v1 | DMT400009189 | CGTCATTCAAAGCCTGTTGTATATTTTTGTGAGGCATAGTGGGAAAGAGATTTTTAAAAC |
Microarray Signals from CUST_5050_PI426222305
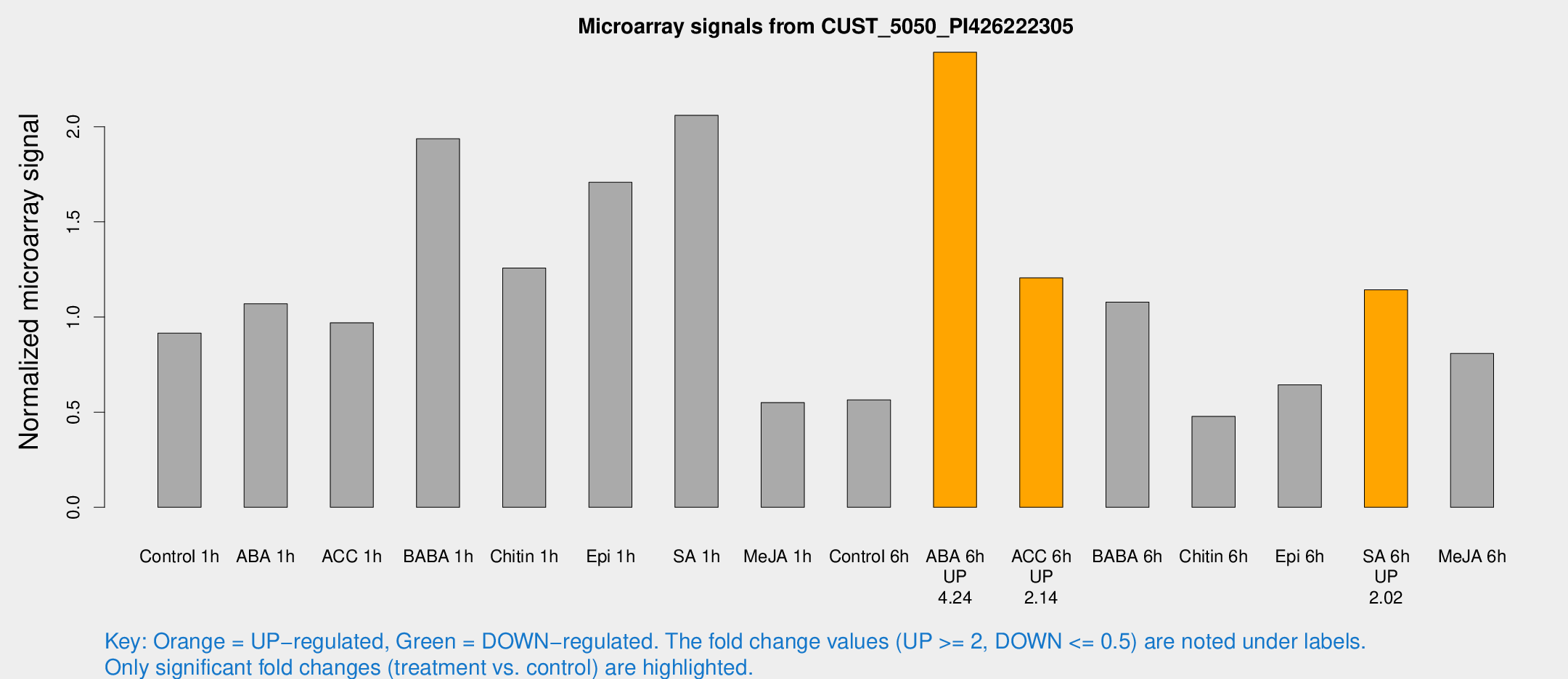
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4727.69 | 347.635 | 0.915066 | 0.0537931 |
| ABA 1h | 6275.75 | 3227.07 | 1.06924 | 0.524515 |
| ACC 1h | 5248.7 | 782.256 | 0.969865 | 0.0854593 |
| BABA 1h | 10931.3 | 3377.33 | 1.93677 | 0.584429 |
| Chitin 1h | 6009.03 | 1149.01 | 1.25733 | 0.142701 |
| Epi 1h | 8263.82 | 2125.65 | 1.70791 | 0.523067 |
| SA 1h | 12190.4 | 4319.84 | 2.0601 | 0.61449 |
| Me-JA 1h | 2412.39 | 525.484 | 0.550573 | 0.07804 |
| Control 6h | 3106.62 | 728.665 | 0.564564 | 0.105398 |
| ABA 6h | 13845.8 | 3205.05 | 2.39149 | 0.46837 |
| ACC 6h | 7139.61 | 469.336 | 1.20585 | 0.107024 |
| BABA 6h | 6825.6 | 2218.89 | 1.07855 | 0.309898 |
| Chitin 6h | 2634.83 | 249.447 | 0.477356 | 0.029839 |
| Epi 6h | 3937.59 | 852.409 | 0.643437 | 0.109465 |
| SA 6h | 5811.15 | 335.608 | 1.14285 | 0.157953 |
| Me-JA 6h | 4461.43 | 1078.66 | 0.808222 | 0.178955 |
Source Transcript PGSC0003DMT400009189 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G17880.1 | +3 | 5e-23 | 93 | 43/62 (69%) | Chaperone DnaJ-domain superfamily protein | chr2:7767176-7767658 REVERSE LENGTH=160 |
