Probe CUST_50326_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50326_PI426222305 | JHI_St_60k_v1 | DMT400041025 | CACCATTTGTAGAAGTTGGGTCATCATCTAGTTATGTTGCAAAAGTTGCGACATTATATT |
All Microarray Probes Designed to Gene DMG400015873
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50326_PI426222305 | JHI_St_60k_v1 | DMT400041025 | CACCATTTGTAGAAGTTGGGTCATCATCTAGTTATGTTGCAAAAGTTGCGACATTATATT |
Microarray Signals from CUST_50326_PI426222305
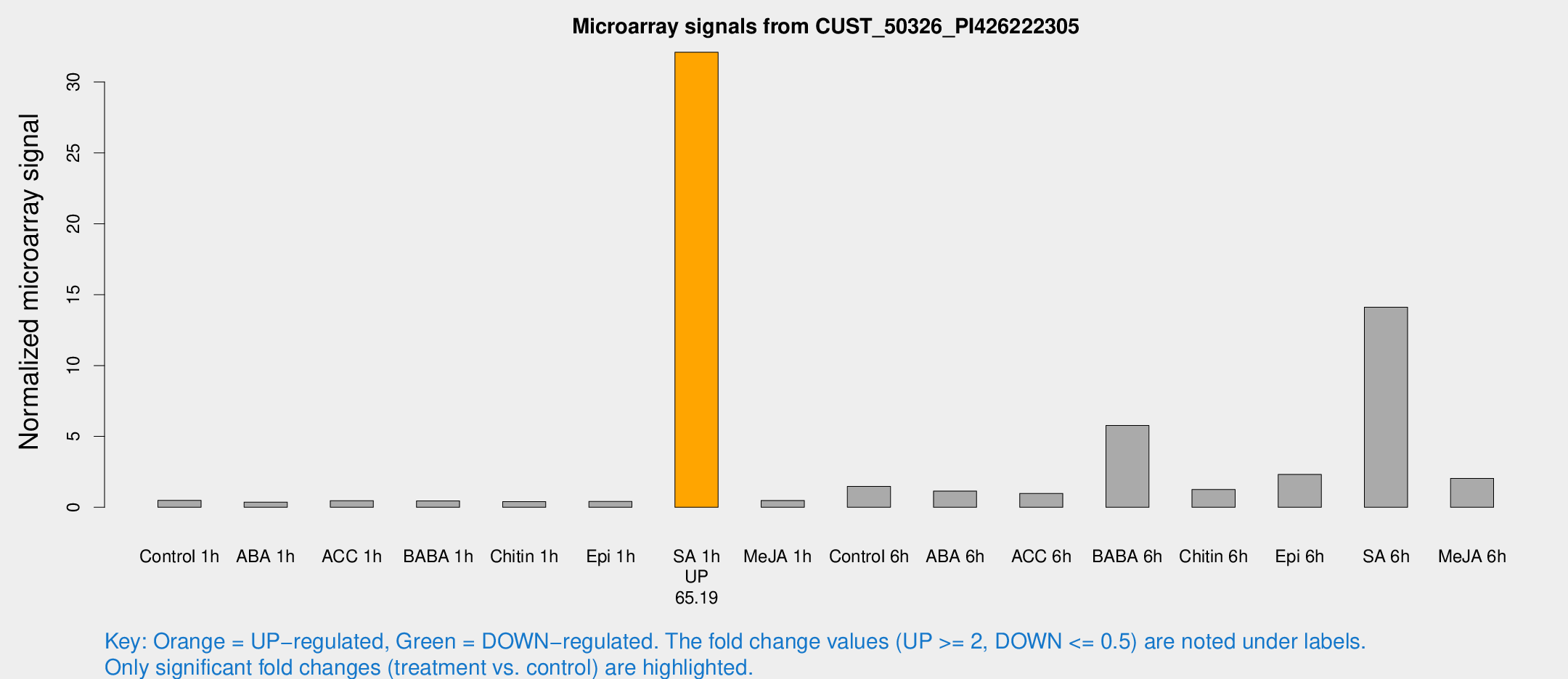
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 15.0087 | 3.21714 | 0.492425 | 0.106191 |
| ABA 1h | 11.2768 | 4.00672 | 0.368486 | 0.167089 |
| ACC 1h | 24.4783 | 17.0735 | 0.470807 | 0.464983 |
| BABA 1h | 17.0472 | 8.01068 | 0.454458 | 0.220187 |
| Chitin 1h | 11.4165 | 3.16209 | 0.403452 | 0.117792 |
| Epi 1h | 13.5504 | 6.50971 | 0.411717 | 0.228295 |
| SA 1h | 1077.47 | 260.175 | 32.1005 | 7.46234 |
| Me-JA 1h | 13.7548 | 4.74885 | 0.483804 | 0.156521 |
| Control 6h | 73.3083 | 37.9702 | 1.47721 | 1.64688 |
| ABA 6h | 41.8587 | 15.2941 | 1.1482 | 0.348039 |
| ACC 6h | 40.6651 | 16.6242 | 0.982197 | 0.674838 |
| BABA 6h | 205.917 | 41.3873 | 5.77005 | 1.48085 |
| Chitin 6h | 40.8886 | 4.26789 | 1.25566 | 0.132589 |
| Epi 6h | 81.3759 | 12.3487 | 2.31429 | 0.260602 |
| SA 6h | 449.837 | 102.78 | 14.1205 | 2.05666 |
| Me-JA 6h | 90.4892 | 50.4141 | 2.04446 | 1.7487 |
Source Transcript PGSC0003DMT400041025 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
