Probe CUST_50208_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50208_PI426222305 | JHI_St_60k_v1 | DMT400080154 | CTTTGTCTGAACTCTGAAGCGAACAATATTTTAAGTTAGCCTAAGAGATGGACTACAAAA |
All Microarray Probes Designed to Gene DMG402031202
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50208_PI426222305 | JHI_St_60k_v1 | DMT400080154 | CTTTGTCTGAACTCTGAAGCGAACAATATTTTAAGTTAGCCTAAGAGATGGACTACAAAA |
Microarray Signals from CUST_50208_PI426222305
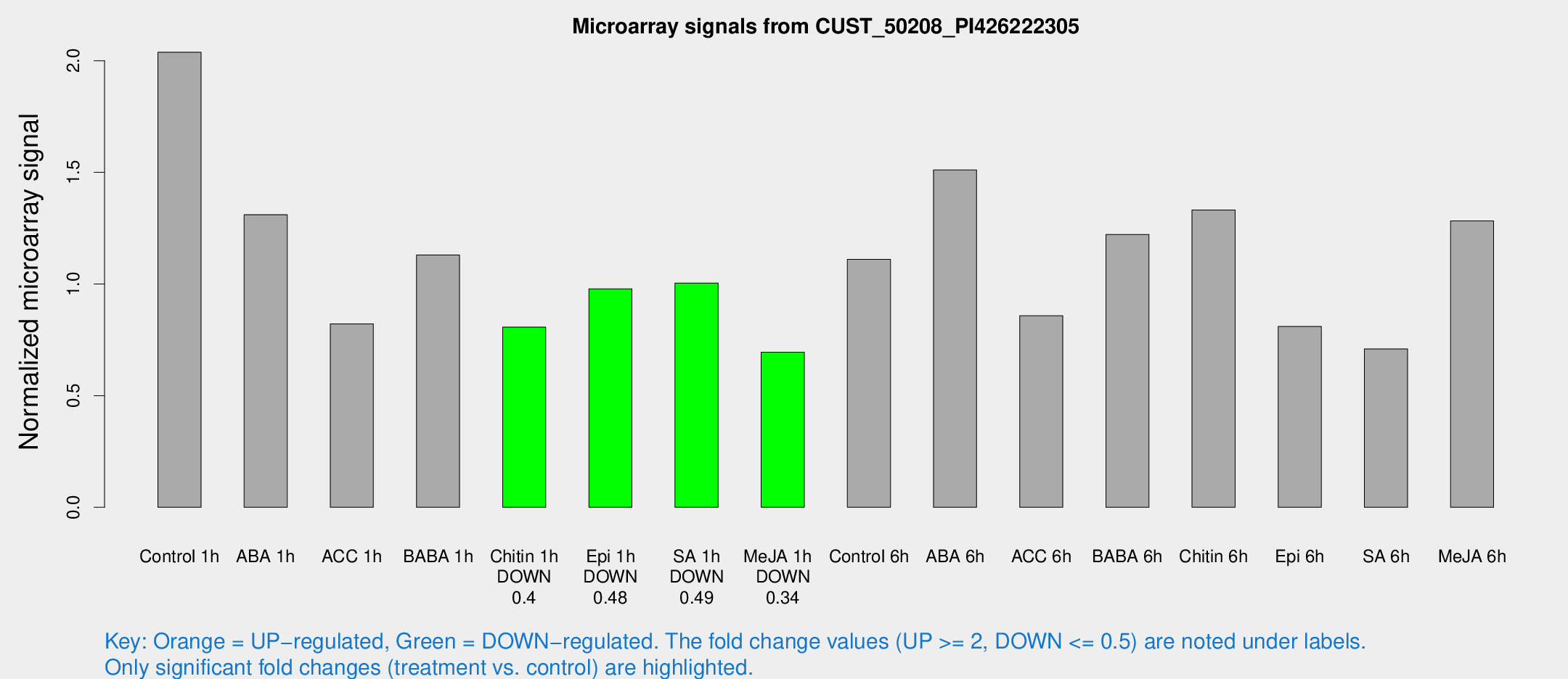
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 185.936 | 21.4489 | 2.03779 | 0.123425 |
| ABA 1h | 111.128 | 27.526 | 1.31035 | 0.314291 |
| ACC 1h | 91.2863 | 33.5946 | 0.821469 | 0.360703 |
| BABA 1h | 105.934 | 28.9666 | 1.12948 | 0.227199 |
| Chitin 1h | 66.0679 | 6.98452 | 0.806758 | 0.0947283 |
| Epi 1h | 77.8916 | 10.844 | 0.978307 | 0.137111 |
| SA 1h | 94.908 | 13.4103 | 1.0044 | 0.114805 |
| Me-JA 1h | 54.1139 | 13.735 | 0.694417 | 0.112382 |
| Control 6h | 106.032 | 22.7781 | 1.11058 | 0.178743 |
| ABA 6h | 156.733 | 39.8846 | 1.51048 | 0.394792 |
| ACC 6h | 90.6481 | 12.954 | 0.857859 | 0.111152 |
| BABA 6h | 129.89 | 27.6332 | 1.22198 | 0.260943 |
| Chitin 6h | 132.33 | 23.417 | 1.33153 | 0.269309 |
| Epi 6h | 88.1387 | 24.5292 | 0.809806 | 0.217705 |
| SA 6h | 65.277 | 11.565 | 0.708858 | 0.219005 |
| Me-JA 6h | 131.525 | 44.71 | 1.28261 | 0.405812 |
Source Transcript PGSC0003DMT400080154 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G56720.1 | +3 | 2e-53 | 196 | 143/345 (41%) | Protein kinase superfamily protein | chr1:21263630-21265559 REVERSE LENGTH=492 |
