Probe CUST_50037_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50037_PI426222305 | JHI_St_60k_v1 | DMT400065441 | GCTCACCACCACTATGCTACAACTATGAGAAGATTTGTAGATTTTATTAATCGAATGATC |
All Microarray Probes Designed to Gene DMG400025449
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50037_PI426222305 | JHI_St_60k_v1 | DMT400065441 | GCTCACCACCACTATGCTACAACTATGAGAAGATTTGTAGATTTTATTAATCGAATGATC |
Microarray Signals from CUST_50037_PI426222305
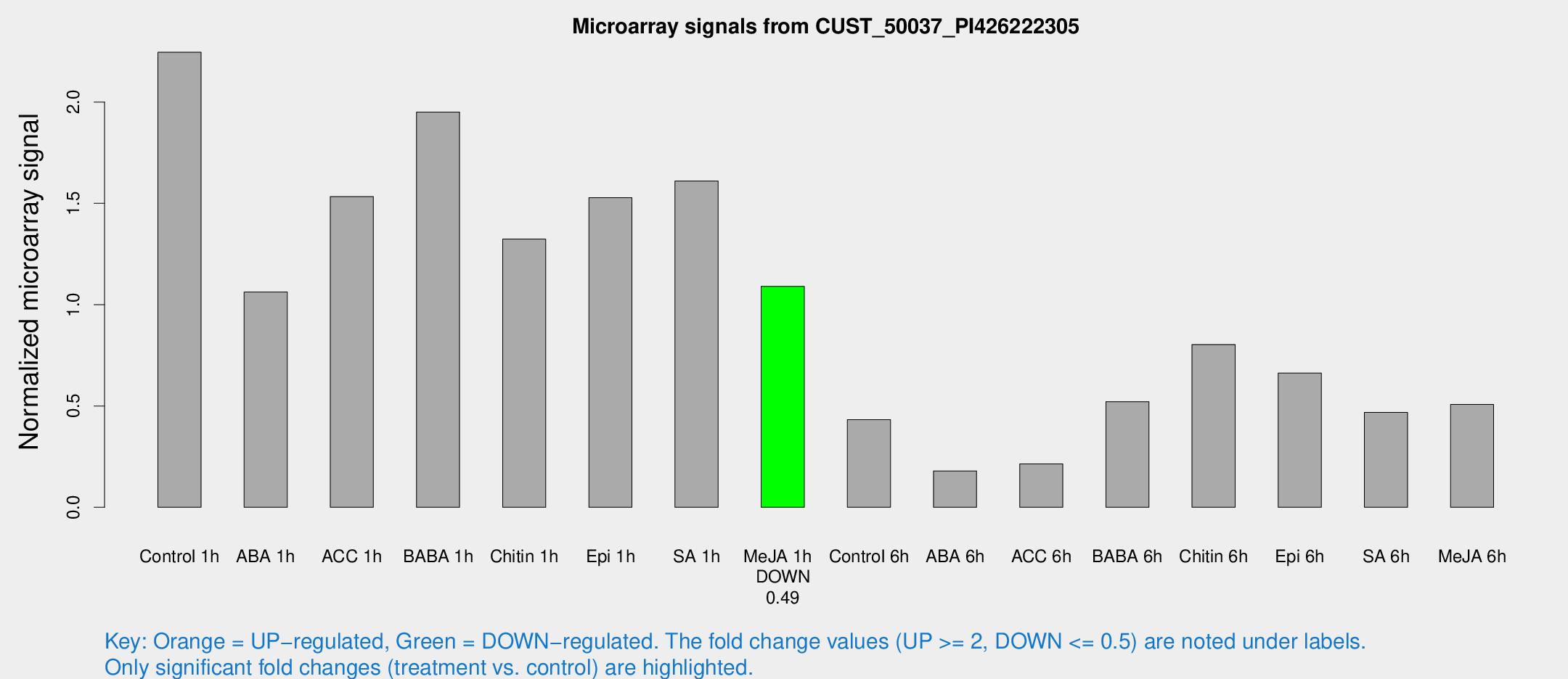
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 90.8135 | 12.1395 | 2.24579 | 0.178466 |
| ABA 1h | 40.0322 | 10.6993 | 1.06268 | 0.238188 |
| ACC 1h | 65.4576 | 14.1704 | 1.53315 | 0.234341 |
| BABA 1h | 74.8485 | 5.506 | 1.9505 | 0.176869 |
| Chitin 1h | 47.6829 | 4.33263 | 1.32399 | 0.183278 |
| Epi 1h | 55.8952 | 13.2523 | 1.52812 | 0.363048 |
| SA 1h | 66.2916 | 5.66508 | 1.61025 | 0.195664 |
| Me-JA 1h | 35.762 | 3.89808 | 1.09023 | 0.119275 |
| Control 6h | 22.2269 | 9.0588 | 0.432903 | 0.236197 |
| ABA 6h | 8.1727 | 3.46638 | 0.179661 | 0.0873401 |
| ACC 6h | 10.4795 | 4.10274 | 0.214033 | 0.0904775 |
| BABA 6h | 23.5035 | 3.92604 | 0.521507 | 0.0883107 |
| Chitin 6h | 34.0181 | 4.14756 | 0.803137 | 0.0980095 |
| Epi 6h | 32.1788 | 8.43492 | 0.662422 | 0.270536 |
| SA 6h | 21.779 | 8.40462 | 0.467939 | 0.241898 |
| Me-JA 6h | 24.1029 | 10.7096 | 0.507411 | 0.199154 |
Source Transcript PGSC0003DMT400065441 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
