Probe CUST_49875_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49875_PI426222305 | JHI_St_60k_v1 | DMT400013096 | GATCTAGAAGAACAACCGGACTTTGATTCCAAGTTGGAACTTCCAACTGATCATGAATAA |
All Microarray Probes Designed to Gene DMG400005113
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49875_PI426222305 | JHI_St_60k_v1 | DMT400013096 | GATCTAGAAGAACAACCGGACTTTGATTCCAAGTTGGAACTTCCAACTGATCATGAATAA |
Microarray Signals from CUST_49875_PI426222305
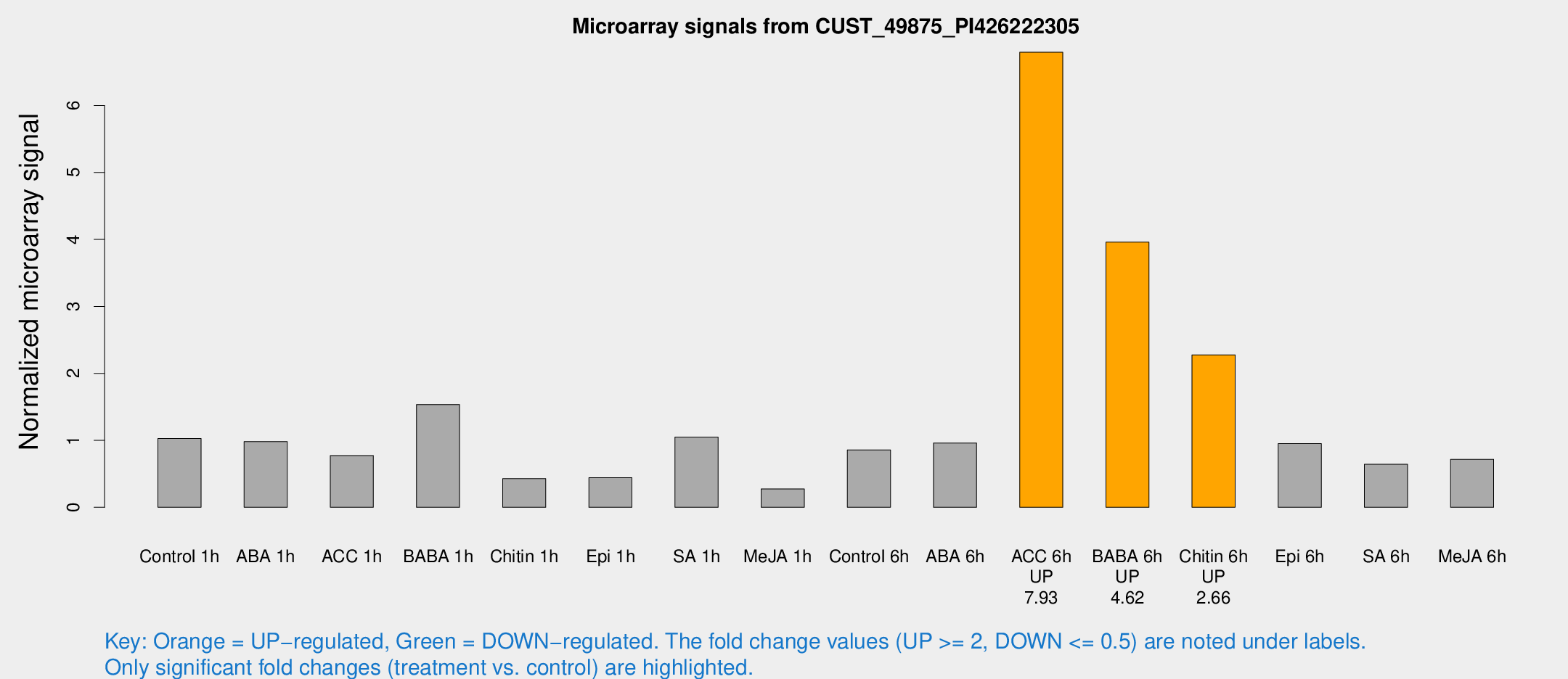
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 175.192 | 95.2569 | 1.02794 | 0.663872 |
| ABA 1h | 182.639 | 84.8546 | 0.980158 | 1.59586 |
| ACC 1h | 167.817 | 79.4669 | 0.772441 | 1.17714 |
| BABA 1h | 218.398 | 74.5272 | 1.53415 | 0.635451 |
| Chitin 1h | 83.6792 | 61.0406 | 0.428182 | 0.413586 |
| Epi 1h | 57.6951 | 23.3758 | 0.441341 | 0.252494 |
| SA 1h | 198.263 | 124.927 | 1.04806 | 0.803117 |
| Me-JA 1h | 39.8233 | 23.4711 | 0.273554 | 0.181541 |
| Control 6h | 113.644 | 30.0765 | 0.85685 | 0.168139 |
| ABA 6h | 446.9 | 291.953 | 0.96095 | 20.3653 |
| ACC 6h | 964.581 | 90.6684 | 6.7965 | 1.58127 |
| BABA 6h | 662.89 | 307.185 | 3.95993 | 1.81819 |
| Chitin 6h | 298.847 | 21.134 | 2.27715 | 0.230175 |
| Epi 6h | 134.228 | 18.5353 | 0.950307 | 0.0737103 |
| SA 6h | 85.0793 | 25.9371 | 0.641897 | 0.198757 |
| Me-JA 6h | 158.735 | 111.012 | 0.715852 | 0.792488 |
Source Transcript PGSC0003DMT400013096 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G33720.1 | +1 | 6e-42 | 138 | 61/92 (66%) | CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein | chr4:16182813-16183304 FORWARD LENGTH=163 |
