Probe CUST_49801_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49801_PI426222305 | JHI_St_60k_v1 | DMT400033777 | AATGGCGCGAGTGCATTGACTTAACAACGTGTGATGCACAAGAAAGTTCACATGTAACGC |
All Microarray Probes Designed to Gene DMG400012977
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49801_PI426222305 | JHI_St_60k_v1 | DMT400033777 | AATGGCGCGAGTGCATTGACTTAACAACGTGTGATGCACAAGAAAGTTCACATGTAACGC |
Microarray Signals from CUST_49801_PI426222305
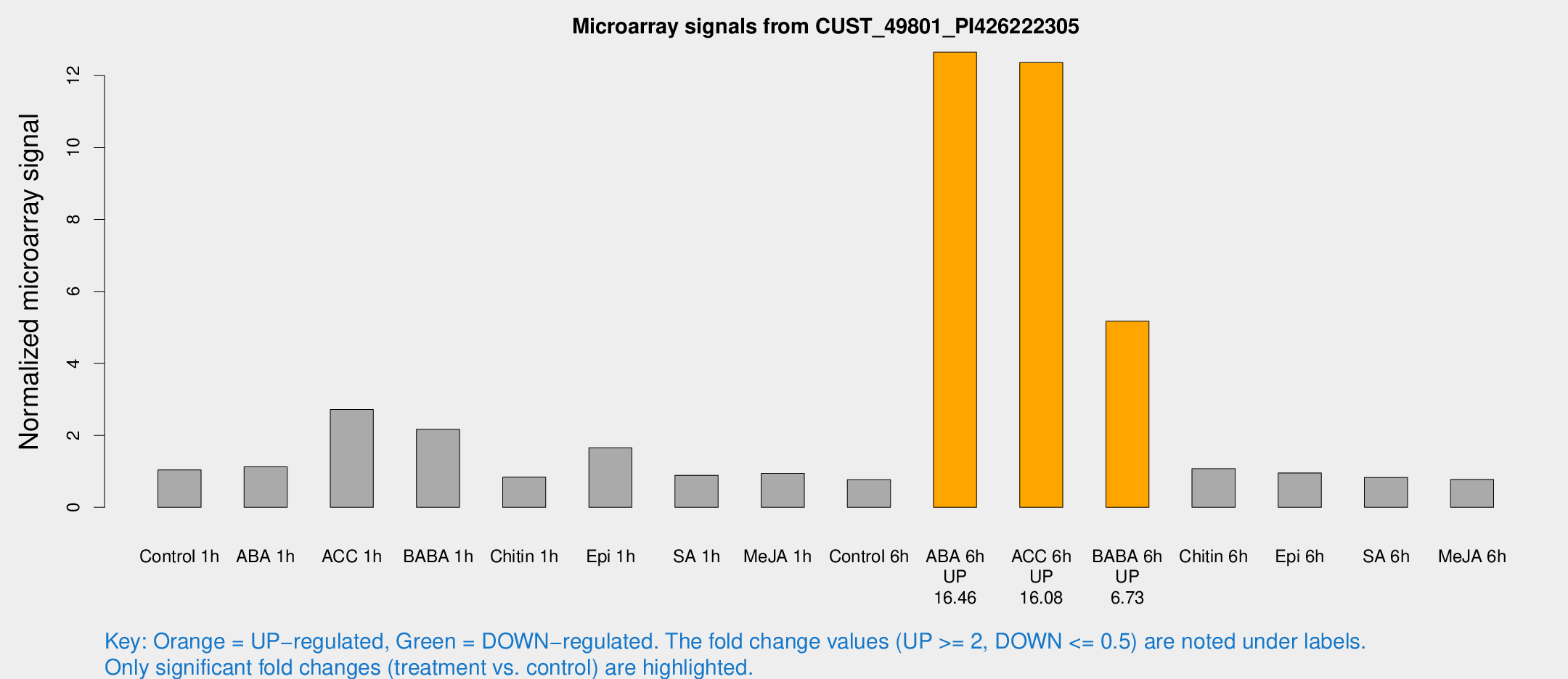
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.84165 | 3.17886 | 1.03803 | 0.459416 |
| ABA 1h | 7.98934 | 3.03458 | 1.12646 | 0.530731 |
| ACC 1h | 26.5156 | 10.2494 | 2.71701 | 1.72043 |
| BABA 1h | 19.6119 | 8.89988 | 2.1667 | 1.11741 |
| Chitin 1h | 5.41939 | 3.15503 | 0.83946 | 0.480084 |
| Epi 1h | 10.4143 | 3.19013 | 1.65634 | 0.510453 |
| SA 1h | 6.95968 | 3.1396 | 0.889702 | 0.44059 |
| Me-JA 1h | 5.61392 | 3.21032 | 0.942977 | 0.538552 |
| Control 6h | 5.59194 | 3.24146 | 0.768688 | 0.445503 |
| ABA 6h | 106.554 | 28.7827 | 12.649 | 3.71813 |
| ACC 6h | 103.936 | 10.2254 | 12.3601 | 1.26206 |
| BABA 6h | 51.6177 | 24.4525 | 5.17146 | 2.38892 |
| Chitin 6h | 9.06034 | 3.59065 | 1.07312 | 0.502529 |
| Epi 6h | 8.19218 | 3.73002 | 0.955028 | 0.471084 |
| SA 6h | 5.93191 | 3.37463 | 0.826202 | 0.470086 |
| Me-JA 6h | 5.59447 | 3.18888 | 0.773319 | 0.440978 |
Source Transcript PGSC0003DMT400033777 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
