Probe CUST_49780_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49780_PI426222305 | JHI_St_60k_v1 | DMT400013922 | GTATTTCTTAGTGGTGTAATGCCTGGTCCAAATATCAGTGCATCAATCGTCTACACTGAG |
All Microarray Probes Designed to Gene DMG400005450
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49780_PI426222305 | JHI_St_60k_v1 | DMT400013922 | GTATTTCTTAGTGGTGTAATGCCTGGTCCAAATATCAGTGCATCAATCGTCTACACTGAG |
Microarray Signals from CUST_49780_PI426222305
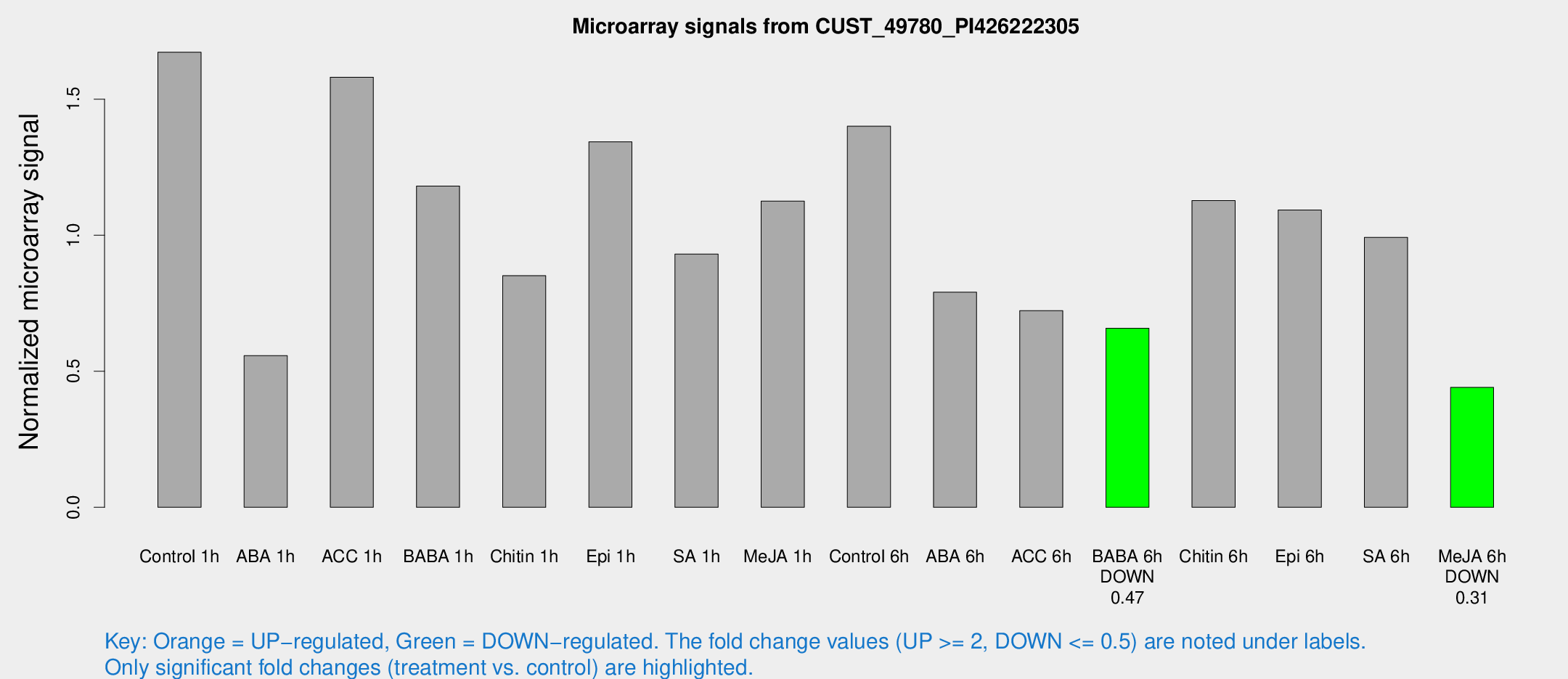
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 25.86 | 7.37201 | 1.67252 | 0.361446 |
| ABA 1h | 7.4556 | 3.288 | 0.557571 | 0.27343 |
| ACC 1h | 24.5937 | 5.68744 | 1.58086 | 0.373598 |
| BABA 1h | 16.9194 | 3.76934 | 1.18073 | 0.319974 |
| Chitin 1h | 13.3694 | 5.80768 | 0.851099 | 0.407719 |
| Epi 1h | 18.3799 | 5.98538 | 1.34319 | 0.38952 |
| SA 1h | 14.1611 | 3.52839 | 0.930726 | 0.250925 |
| Me-JA 1h | 14.5661 | 4.12024 | 1.12559 | 0.503124 |
| Control 6h | 20.582 | 3.73177 | 1.40056 | 0.271652 |
| ABA 6h | 15.2453 | 7.44322 | 0.790512 | 0.499687 |
| ACC 6h | 13.2173 | 4.18991 | 0.722788 | 0.272321 |
| BABA 6h | 10.5866 | 3.96363 | 0.658223 | 0.245941 |
| Chitin 6h | 17.9678 | 4.02833 | 1.12704 | 0.275826 |
| Epi 6h | 19.8548 | 5.90031 | 1.09261 | 0.546977 |
| SA 6h | 14.195 | 3.83751 | 0.992074 | 0.272936 |
| Me-JA 6h | 6.3179 | 3.50526 | 0.440459 | 0.245368 |
Source Transcript PGSC0003DMT400013922 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G50440.1 | +1 | 8e-68 | 215 | 113/267 (42%) | methyl esterase 10 | chr3:18717392-18718435 REVERSE LENGTH=288 |
