Probe CUST_49766_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49766_PI426222305 | JHI_St_60k_v1 | DMT400035859 | GATCAATTCATAAATTCAAGCTTGTTTGGTGGAGACTTGAAAGTTGGAGTGGAAAAGGGA |
All Microarray Probes Designed to Gene DMG400013807
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49766_PI426222305 | JHI_St_60k_v1 | DMT400035859 | GATCAATTCATAAATTCAAGCTTGTTTGGTGGAGACTTGAAAGTTGGAGTGGAAAAGGGA |
Microarray Signals from CUST_49766_PI426222305
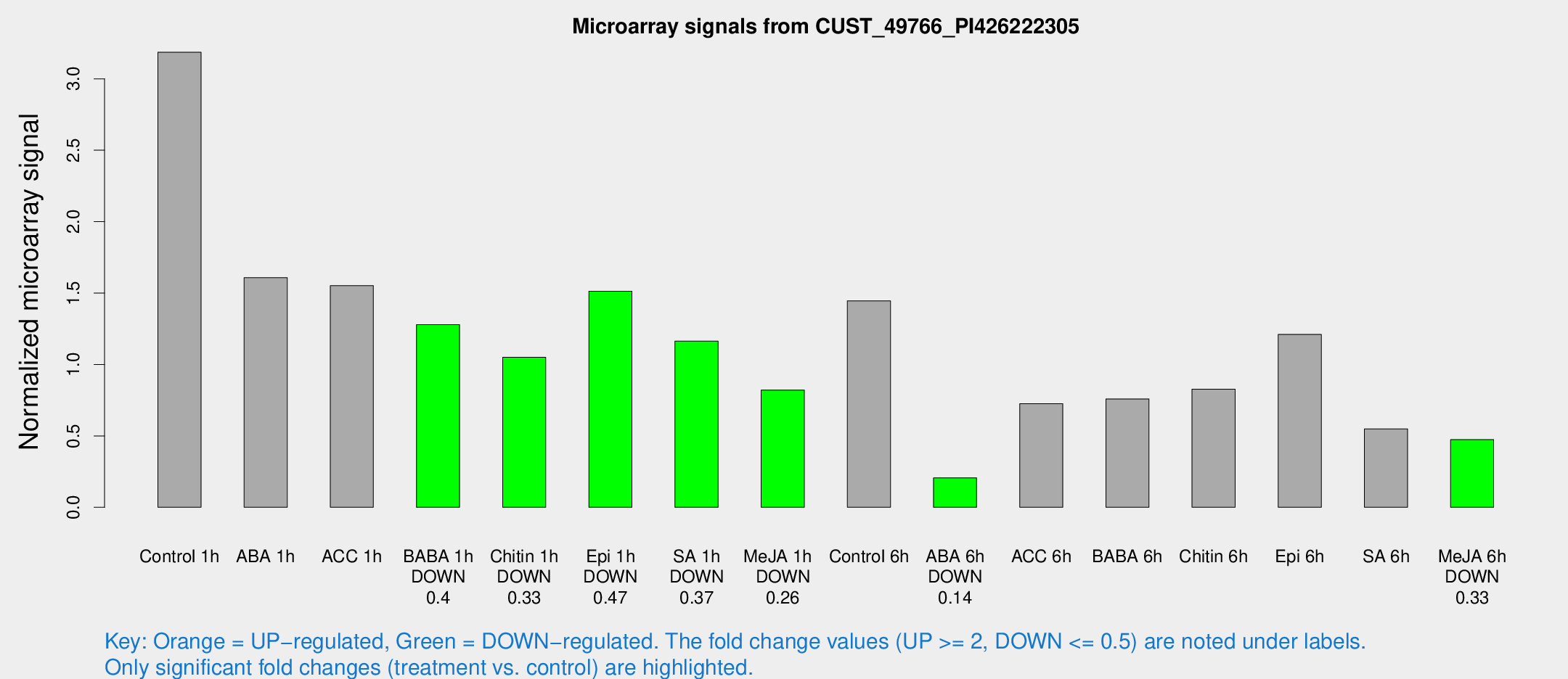
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 900.376 | 191.203 | 3.18631 | 0.473043 |
| ABA 1h | 388.491 | 40.6296 | 1.60779 | 0.143582 |
| ACC 1h | 467.76 | 117.431 | 1.55193 | 0.367492 |
| BABA 1h | 356.926 | 83.089 | 1.27933 | 0.224644 |
| Chitin 1h | 259.715 | 34.4797 | 1.05046 | 0.0618544 |
| Epi 1h | 357.074 | 33.9208 | 1.51278 | 0.126355 |
| SA 1h | 334.984 | 60.6898 | 1.16323 | 0.187098 |
| Me-JA 1h | 182.037 | 14.1629 | 0.820732 | 0.0548619 |
| Control 6h | 413.52 | 101.574 | 1.44607 | 0.305824 |
| ABA 6h | 59.8332 | 5.97631 | 0.206276 | 0.0329414 |
| ACC 6h | 233.467 | 47.2144 | 0.725727 | 0.0483497 |
| BABA 6h | 235.419 | 38.4268 | 0.758825 | 0.108914 |
| Chitin 6h | 240.105 | 23.2756 | 0.826703 | 0.0651117 |
| Epi 6h | 369.404 | 21.6602 | 1.21092 | 0.123388 |
| SA 6h | 198.48 | 85.3968 | 0.549394 | 0.33621 |
| Me-JA 6h | 139.498 | 43.5133 | 0.473928 | 0.122536 |
Source Transcript PGSC0003DMT400035859 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G27570.1 | +1 | 9e-52 | 174 | 77/145 (53%) | UDP-Glycosyltransferase superfamily protein | chr4:13763657-13765018 REVERSE LENGTH=453 |
