Probe CUST_49749_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49749_PI426222305 | JHI_St_60k_v1 | DMT400035873 | CAGAATATGGGCAATGGCTATAGCAATAATGTTAAGTGTTACAATGATGAGAAGCAATCA |
All Microarray Probes Designed to Gene DMG400013810
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49746_PI426222305 | JHI_St_60k_v1 | DMT400035872 | GAAATGGAGGCACGATTATCGGGATGTATCCGAAAGTGGGATGCGATTATCAGGATGTTT |
| CUST_49749_PI426222305 | JHI_St_60k_v1 | DMT400035873 | CAGAATATGGGCAATGGCTATAGCAATAATGTTAAGTGTTACAATGATGAGAAGCAATCA |
Microarray Signals from CUST_49749_PI426222305
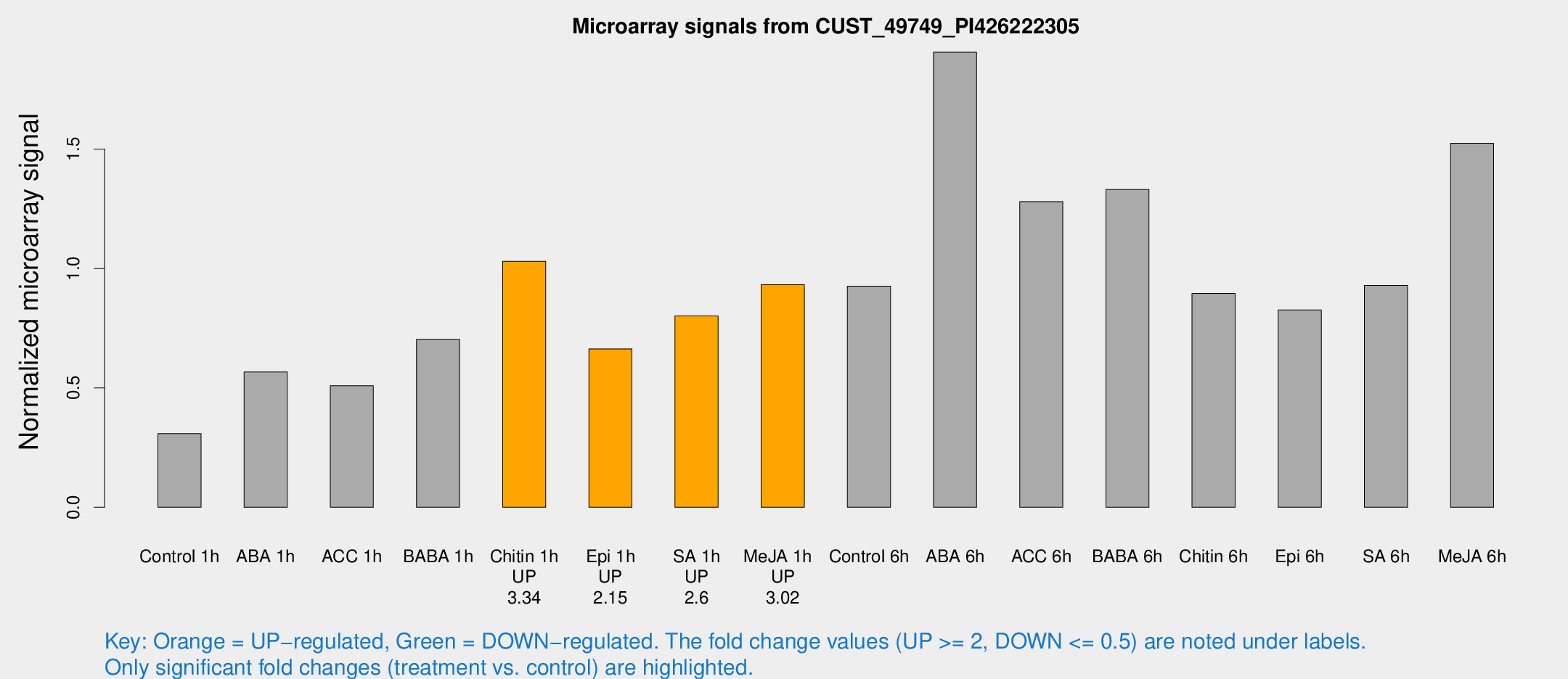
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 162.717 | 9.95933 | 0.3087 | 0.0281602 |
| ABA 1h | 267.966 | 33.3085 | 0.566764 | 0.0361603 |
| ACC 1h | 289.372 | 59.9829 | 0.509376 | 0.162965 |
| BABA 1h | 425.344 | 142.534 | 0.703353 | 0.278538 |
| Chitin 1h | 489.932 | 36.4206 | 1.03031 | 0.0598661 |
| Epi 1h | 302.099 | 17.7326 | 0.663493 | 0.0389354 |
| SA 1h | 462.754 | 125.484 | 0.801223 | 0.162042 |
| Me-JA 1h | 422.254 | 89.552 | 0.931872 | 0.156177 |
| Control 6h | 511.483 | 103.232 | 0.925738 | 0.1315 |
| ABA 6h | 1176.93 | 352.432 | 1.9057 | 0.541941 |
| ACC 6h | 807.664 | 177.533 | 1.28002 | 0.0918462 |
| BABA 6h | 786.196 | 48.7718 | 1.33057 | 0.0770842 |
| Chitin 6h | 514.553 | 83.6583 | 0.895814 | 0.128866 |
| Epi 6h | 544.002 | 149.98 | 0.826269 | 0.24565 |
| SA 6h | 531.072 | 144.577 | 0.928761 | 0.19997 |
| Me-JA 6h | 862.007 | 210.094 | 1.52415 | 0.349661 |
Source Transcript PGSC0003DMT400035873 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G21460.1 | +3 | 1e-76 | 238 | 137/241 (57%) | Nodulin MtN3 family protein | chr1:7512030-7513281 REVERSE LENGTH=247 |
