Probe CUST_49042_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49042_PI426222305 | JHI_St_60k_v1 | DMT400044014 | AAGAGGAGGATTTTAAAGGATCTTCTGTTAAAGCATTGGATGATTTCATATCTACTCTCC |
All Microarray Probes Designed to Gene DMG402017077
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49042_PI426222305 | JHI_St_60k_v1 | DMT400044014 | AAGAGGAGGATTTTAAAGGATCTTCTGTTAAAGCATTGGATGATTTCATATCTACTCTCC |
Microarray Signals from CUST_49042_PI426222305
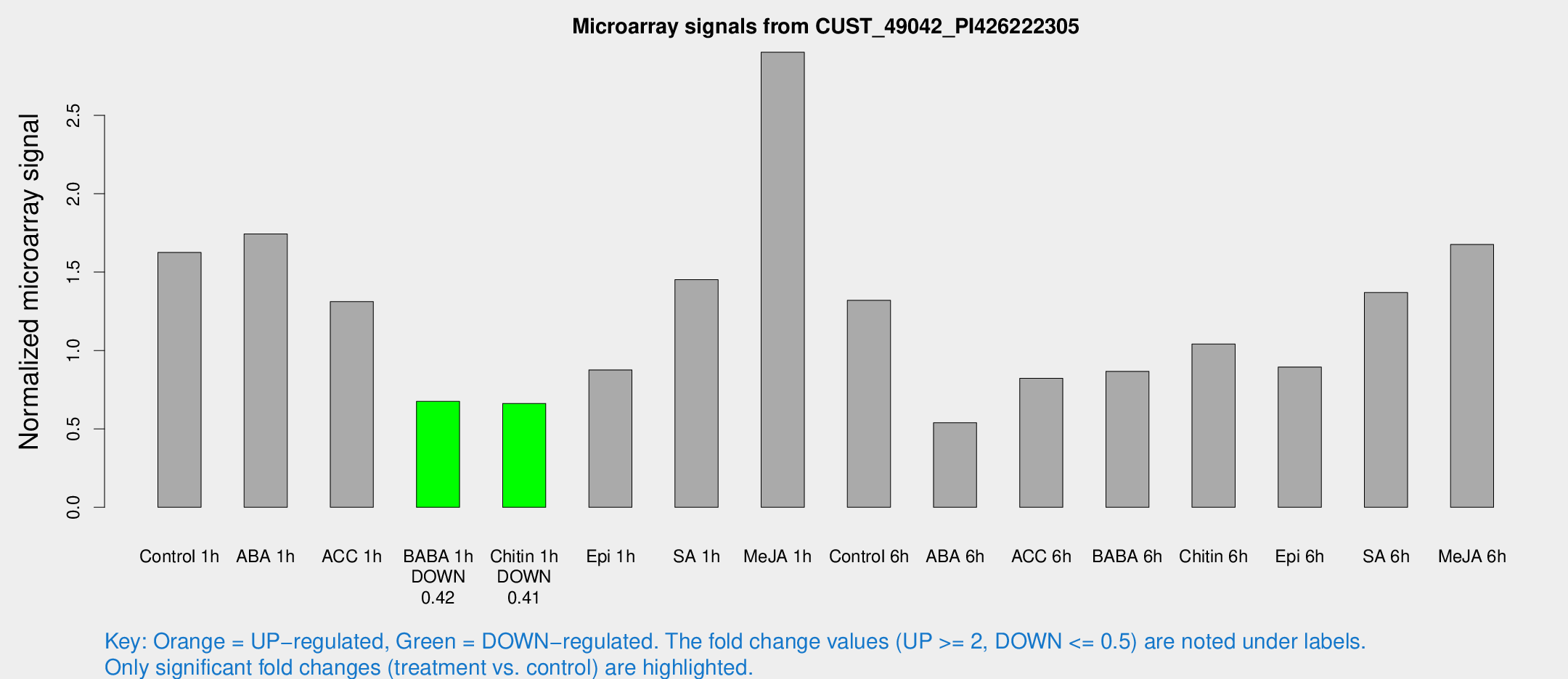
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 467.016 | 105.211 | 1.62503 | 0.24857 |
| ABA 1h | 424.497 | 24.8269 | 1.74384 | 0.101717 |
| ACC 1h | 401.347 | 100.555 | 1.31157 | 0.309658 |
| BABA 1h | 180.276 | 16.9792 | 0.675263 | 0.0416541 |
| Chitin 1h | 164.466 | 13.7371 | 0.662193 | 0.0659829 |
| Epi 1h | 208.089 | 12.5246 | 0.876299 | 0.0527425 |
| SA 1h | 424.935 | 84.6663 | 1.45177 | 0.328056 |
| Me-JA 1h | 650.281 | 37.7442 | 2.90226 | 0.198792 |
| Control 6h | 380.979 | 78.4167 | 1.32004 | 0.193911 |
| ABA 6h | 157.659 | 9.90801 | 0.539595 | 0.0467718 |
| ACC 6h | 263.201 | 33.2693 | 0.822563 | 0.0838137 |
| BABA 6h | 267.962 | 20.6589 | 0.867075 | 0.0968509 |
| Chitin 6h | 304.693 | 18.1047 | 1.0414 | 0.0637375 |
| Epi 6h | 286.157 | 52.5295 | 0.89488 | 0.24317 |
| SA 6h | 437.673 | 143.404 | 1.36999 | 0.481901 |
| Me-JA 6h | 460.534 | 34.1639 | 1.67589 | 0.206248 |
Source Transcript PGSC0003DMT400044014 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G12890.1 | +1 | 5e-47 | 162 | 87/187 (47%) | UDP-Glycosyltransferase superfamily protein | chr5:4069658-4071124 REVERSE LENGTH=488 |
