Probe CUST_48737_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48737_PI426222305 | JHI_St_60k_v1 | DMT400048249 | TCGTATTTGACGGCCAAGATGAAAGGAGGTTTCCCTGTTACTATTGAGGAAAGGATGTAG |
All Microarray Probes Designed to Gene DMG400018744
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48737_PI426222305 | JHI_St_60k_v1 | DMT400048249 | TCGTATTTGACGGCCAAGATGAAAGGAGGTTTCCCTGTTACTATTGAGGAAAGGATGTAG |
Microarray Signals from CUST_48737_PI426222305
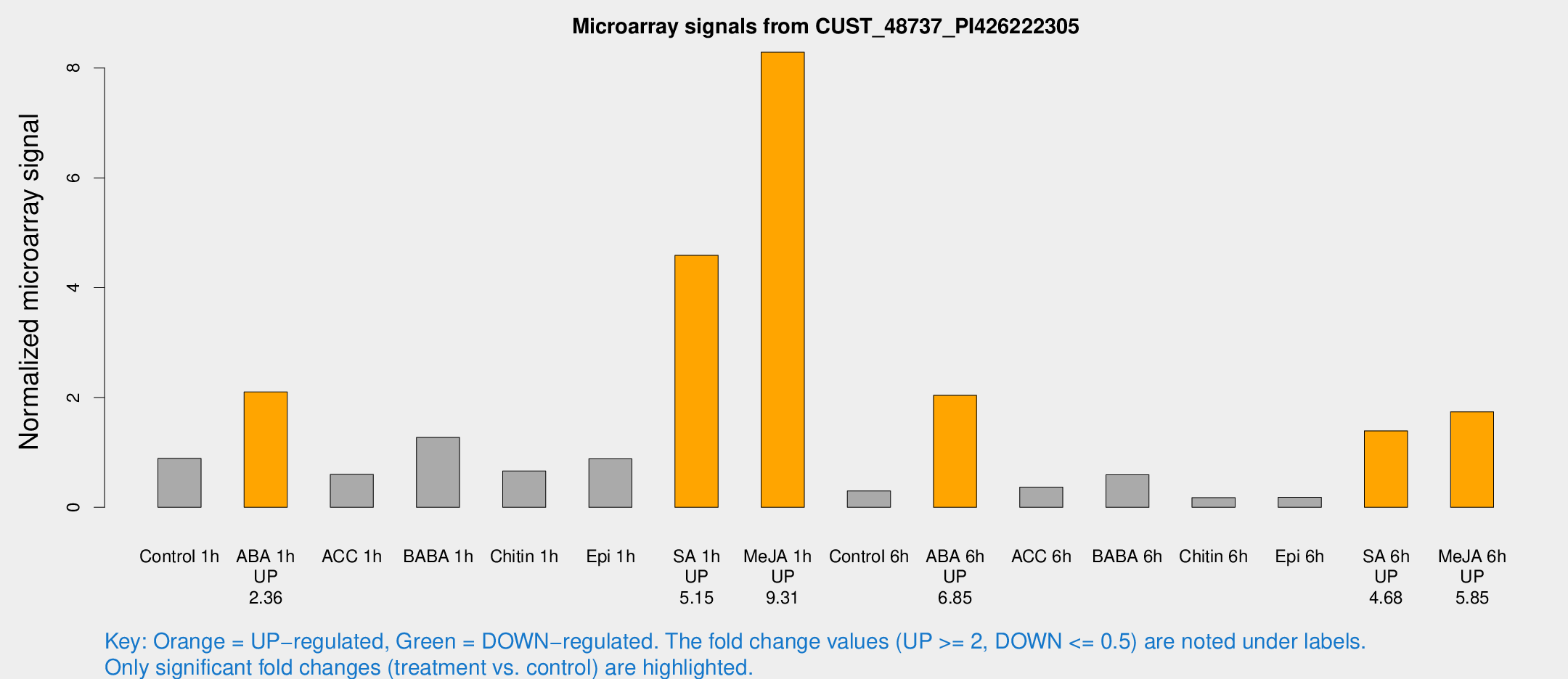
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 932.732 | 83.4712 | 0.890479 | 0.0586765 |
| ABA 1h | 1992.45 | 326.488 | 2.10152 | 0.25823 |
| ACC 1h | 776.544 | 332.311 | 0.597708 | 0.263799 |
| BABA 1h | 1287.61 | 127.794 | 1.27335 | 0.13482 |
| Chitin 1h | 645.141 | 125.897 | 0.661034 | 0.113772 |
| Epi 1h | 822.764 | 162.067 | 0.882697 | 0.183802 |
| SA 1h | 5343.31 | 1445.65 | 4.59 | 1.29409 |
| Me-JA 1h | 7248.07 | 1203.67 | 8.28729 | 0.797007 |
| Control 6h | 359.719 | 126.385 | 0.297478 | 0.0998349 |
| ABA 6h | 2528.1 | 836.315 | 2.03897 | 0.626021 |
| ACC 6h | 442.031 | 40.604 | 0.367774 | 0.0759663 |
| BABA 6h | 811.315 | 285.573 | 0.593833 | 0.312145 |
| Chitin 6h | 198.238 | 30.3215 | 0.175274 | 0.0256314 |
| Epi 6h | 218.264 | 36.4679 | 0.181501 | 0.0320163 |
| SA 6h | 1487.73 | 297.914 | 1.39281 | 0.188648 |
| Me-JA 6h | 1968.21 | 616.614 | 1.73977 | 0.706368 |
Source Transcript PGSC0003DMT400048249 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G63450.1 | +1 | 8e-149 | 439 | 222/469 (47%) | cytochrome P450, family 94, subfamily B, polypeptide 1 | chr5:25408987-25410519 REVERSE LENGTH=510 |
