Probe CUST_48665_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48665_PI426222305 | JHI_St_60k_v1 | DMT400064975 | GCTCGTTGGCTTAAAAAATTCCTTTCTAGTAGAGACATTGTTTCCAGATCTGATTGTTAA |
All Microarray Probes Designed to Gene DMG400025239
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48631_PI426222305 | JHI_St_60k_v1 | DMT400064978 | ATCATTATCACCCCCTCAACGATGTTCAGATGAATTATAACGACGAATGCTACTATAAGG |
| CUST_48665_PI426222305 | JHI_St_60k_v1 | DMT400064975 | GCTCGTTGGCTTAAAAAATTCCTTTCTAGTAGAGACATTGTTTCCAGATCTGATTGTTAA |
| CUST_48663_PI426222305 | JHI_St_60k_v1 | DMT400064976 | GTAGGTATAAAAAATGGAATACCCATCGGCAAACGGACAGGATCGAAAGATGATTTCGTC |
Microarray Signals from CUST_48665_PI426222305
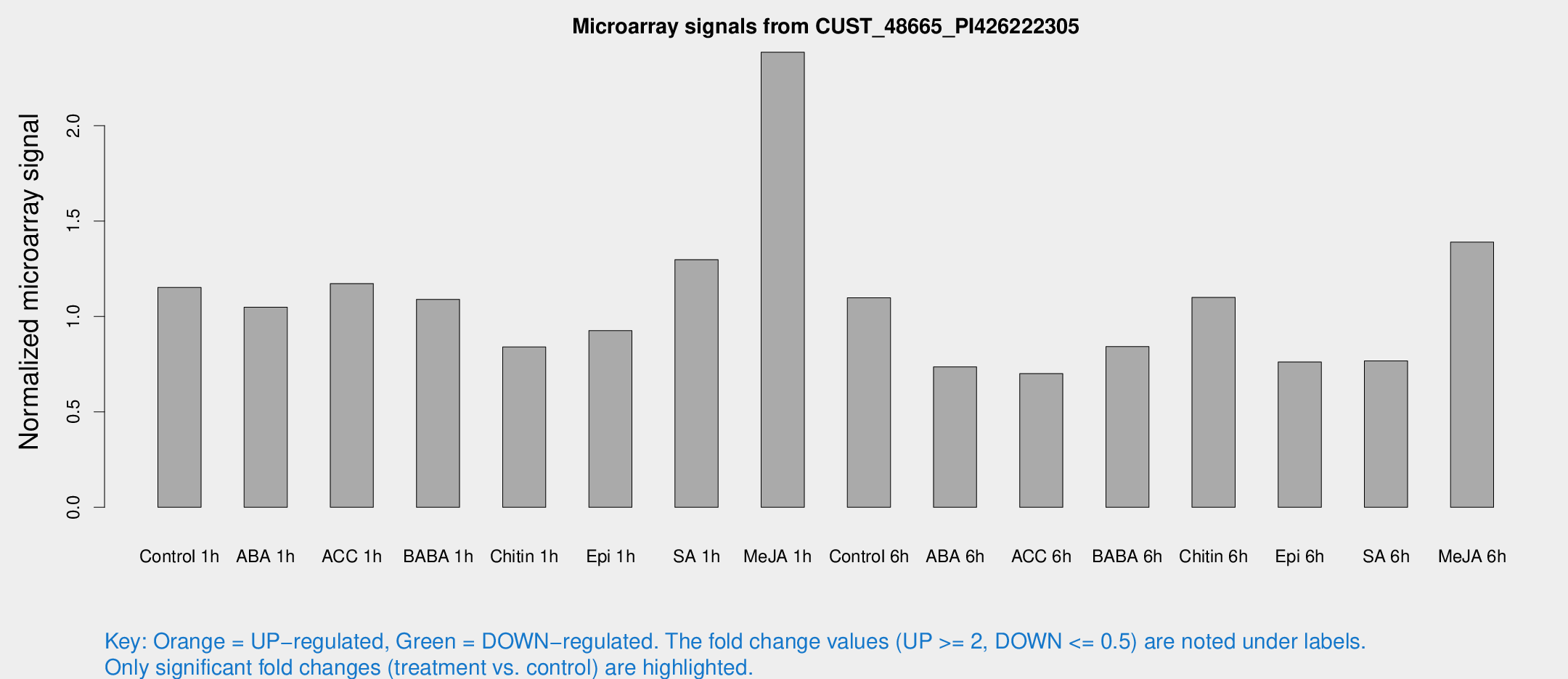
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1188.2 | 68.8817 | 1.15232 | 0.0974812 |
| ABA 1h | 998.373 | 225.907 | 1.04805 | 0.161574 |
| ACC 1h | 1380.08 | 386.874 | 1.17219 | 0.344344 |
| BABA 1h | 1089.57 | 105.906 | 1.08956 | 0.150902 |
| Chitin 1h | 777.58 | 45.0639 | 0.840286 | 0.0786539 |
| Epi 1h | 849.745 | 140.831 | 0.925921 | 0.161575 |
| SA 1h | 1582.77 | 558.47 | 1.29762 | 0.650347 |
| Me-JA 1h | 2011.67 | 148.522 | 2.38466 | 0.304729 |
| Control 6h | 1202.21 | 267.988 | 1.09785 | 0.181997 |
| ABA 6h | 816.363 | 101.112 | 0.735919 | 0.133324 |
| ACC 6h | 846.119 | 134.344 | 0.700546 | 0.0455649 |
| BABA 6h | 983.568 | 124.033 | 0.842097 | 0.107066 |
| Chitin 6h | 1205.49 | 69.8394 | 1.09953 | 0.0681753 |
| Epi 6h | 898.169 | 117.102 | 0.761287 | 0.165635 |
| SA 6h | 789.785 | 88.7934 | 0.767001 | 0.0453493 |
| Me-JA 6h | 1506.26 | 340.955 | 1.38955 | 0.227791 |
Source Transcript PGSC0003DMT400064975 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G44640.1 | +1 | 6e-96 | 297 | 142/292 (49%) | beta glucosidase 13 | chr5:18011146-18012669 FORWARD LENGTH=507 |
