Probe CUST_48457_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48457_PI426222305 | JHI_St_60k_v1 | DMT400055808 | GCACAGCCTATATGGGTGATAGAATGTTTGTAACATAATTGAATTTGCATTGATACCTAC |
All Microarray Probes Designed to Gene DMG400021676
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48430_PI426222305 | JHI_St_60k_v1 | DMT400055809 | TGAAGATGAGCCTGTTCCCTTATCTCAAGTCAAGATAGGACAAGAGTATGAAATTGTCCT |
| CUST_48457_PI426222305 | JHI_St_60k_v1 | DMT400055808 | GCACAGCCTATATGGGTGATAGAATGTTTGTAACATAATTGAATTTGCATTGATACCTAC |
Microarray Signals from CUST_48457_PI426222305
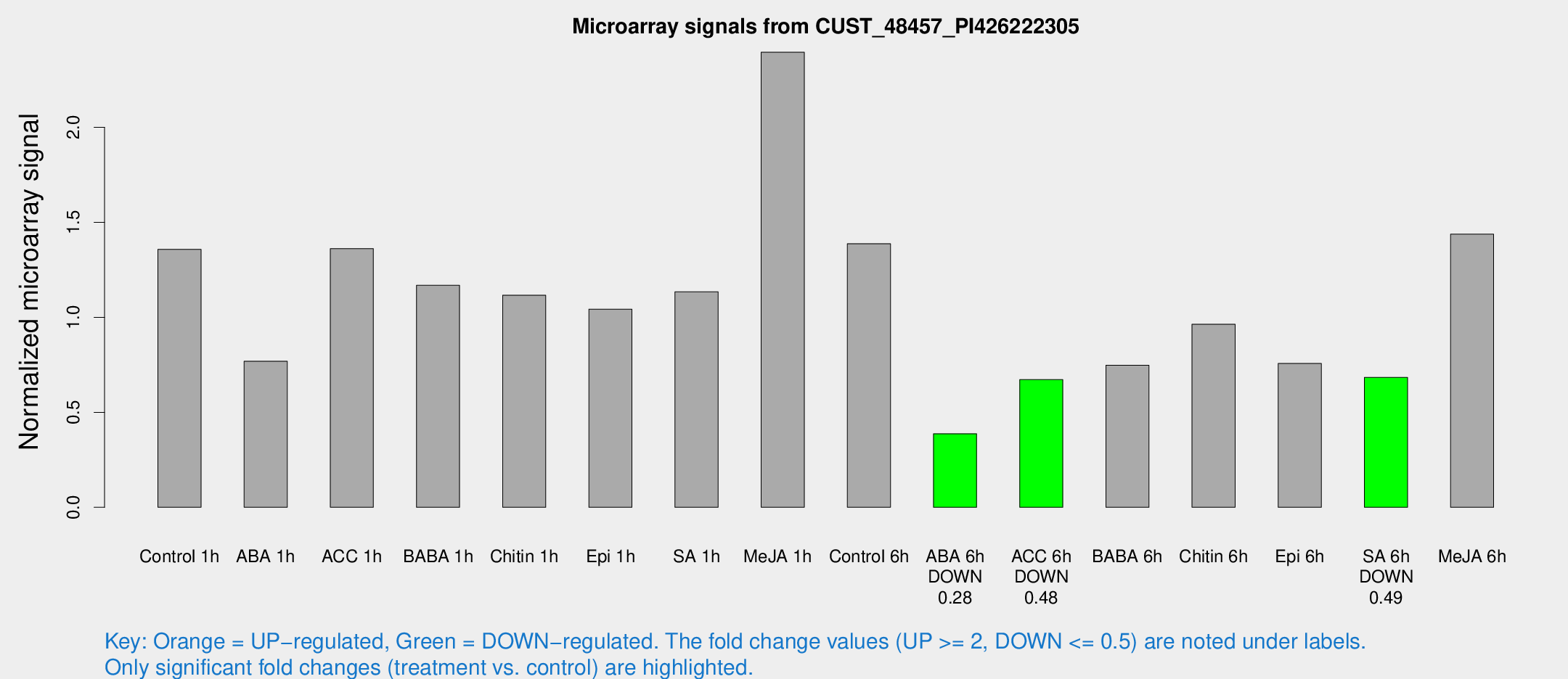
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5649.43 | 1402.44 | 1.35798 | 0.273917 |
| ABA 1h | 2711.96 | 326.951 | 0.76882 | 0.0520299 |
| ACC 1h | 6014.11 | 1846.45 | 1.36164 | 0.35707 |
| BABA 1h | 4842.8 | 1265.42 | 1.16839 | 0.2617 |
| Chitin 1h | 3960.14 | 276.761 | 1.11635 | 0.0721709 |
| Epi 1h | 3578.14 | 342.285 | 1.0429 | 0.0873449 |
| SA 1h | 4629.85 | 531.491 | 1.134 | 0.137831 |
| Me-JA 1h | 7910.97 | 1317 | 2.39528 | 0.225968 |
| Control 6h | 5608.31 | 866.445 | 1.38755 | 0.100891 |
| ABA 6h | 1632.84 | 164.812 | 0.387251 | 0.0339834 |
| ACC 6h | 3158.44 | 628.868 | 0.671846 | 0.0779185 |
| BABA 6h | 3342.29 | 445.952 | 0.747829 | 0.0875678 |
| Chitin 6h | 4066.85 | 413.882 | 0.963631 | 0.109594 |
| Epi 6h | 3494.98 | 687.202 | 0.75695 | 0.207405 |
| SA 6h | 2736.7 | 445.39 | 0.683816 | 0.0421442 |
| Me-JA 6h | 5640.73 | 338.62 | 1.43766 | 0.0830066 |
Source Transcript PGSC0003DMT400055808 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G03400.1 | +1 | 4e-79 | 251 | 121/180 (67%) | Auxin-responsive GH3 family protein | chr4:1497675-1499729 REVERSE LENGTH=591 |
