Probe CUST_48430_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48430_PI426222305 | JHI_St_60k_v1 | DMT400055809 | TGAAGATGAGCCTGTTCCCTTATCTCAAGTCAAGATAGGACAAGAGTATGAAATTGTCCT |
All Microarray Probes Designed to Gene DMG400021676
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48457_PI426222305 | JHI_St_60k_v1 | DMT400055808 | GCACAGCCTATATGGGTGATAGAATGTTTGTAACATAATTGAATTTGCATTGATACCTAC |
| CUST_48430_PI426222305 | JHI_St_60k_v1 | DMT400055809 | TGAAGATGAGCCTGTTCCCTTATCTCAAGTCAAGATAGGACAAGAGTATGAAATTGTCCT |
Microarray Signals from CUST_48430_PI426222305
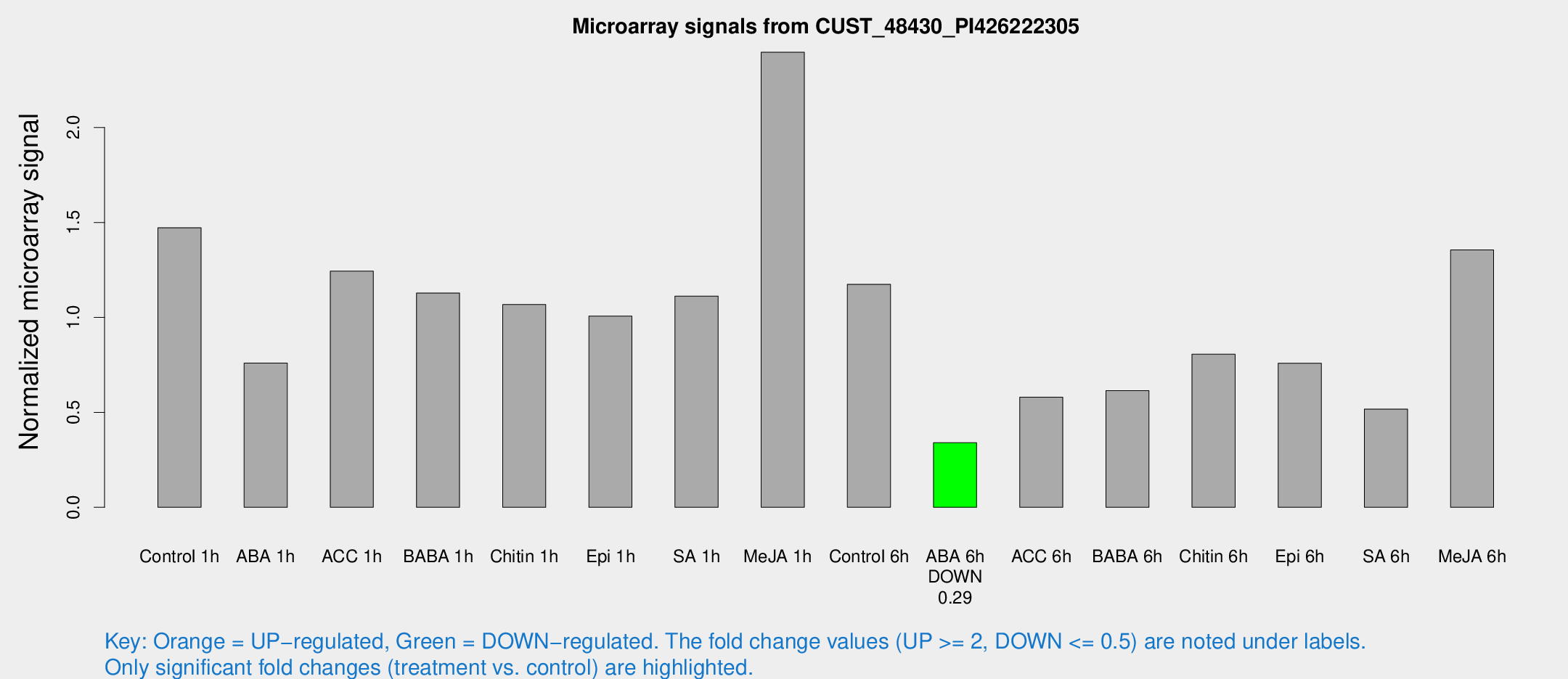
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3411.91 | 941.059 | 1.47212 | 0.306927 |
| ABA 1h | 1501 | 260.439 | 0.759186 | 0.0897741 |
| ACC 1h | 3061.3 | 934.124 | 1.24362 | 0.346551 |
| BABA 1h | 2647.62 | 764.375 | 1.12837 | 0.315271 |
| Chitin 1h | 2089.79 | 161.34 | 1.06807 | 0.0616853 |
| Epi 1h | 1911.58 | 217.969 | 1.00681 | 0.101231 |
| SA 1h | 2509.99 | 308.686 | 1.1125 | 0.119796 |
| Me-JA 1h | 4467.22 | 942.578 | 2.39661 | 0.405214 |
| Control 6h | 2760.06 | 704.18 | 1.17446 | 0.256253 |
| ABA 6h | 784.951 | 45.5884 | 0.340182 | 0.0196934 |
| ACC 6h | 1500.79 | 302.549 | 0.580052 | 0.0544772 |
| BABA 6h | 1529.85 | 268.535 | 0.614051 | 0.101269 |
| Chitin 6h | 1882.35 | 228.217 | 0.806171 | 0.0968653 |
| Epi 6h | 1966.51 | 484.045 | 0.758923 | 0.225848 |
| SA 6h | 1340.81 | 530.647 | 0.517103 | 0.200317 |
| Me-JA 6h | 2947.56 | 295.912 | 1.35519 | 0.174984 |
Source Transcript PGSC0003DMT400055809 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G03400.1 | +2 | 0.0 | 823 | 398/590 (67%) | Auxin-responsive GH3 family protein | chr4:1497675-1499729 REVERSE LENGTH=591 |
