Probe CUST_48030_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48030_PI426222305 | JHI_St_60k_v1 | DMT400021782 | TTACGACCTCCAGCAAGCTAAAGTAAGTATTAAGAACATGGTAGAAGAAGAAGAAGGGAG |
All Microarray Probes Designed to Gene DMG400008453
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48030_PI426222305 | JHI_St_60k_v1 | DMT400021782 | TTACGACCTCCAGCAAGCTAAAGTAAGTATTAAGAACATGGTAGAAGAAGAAGAAGGGAG |
Microarray Signals from CUST_48030_PI426222305
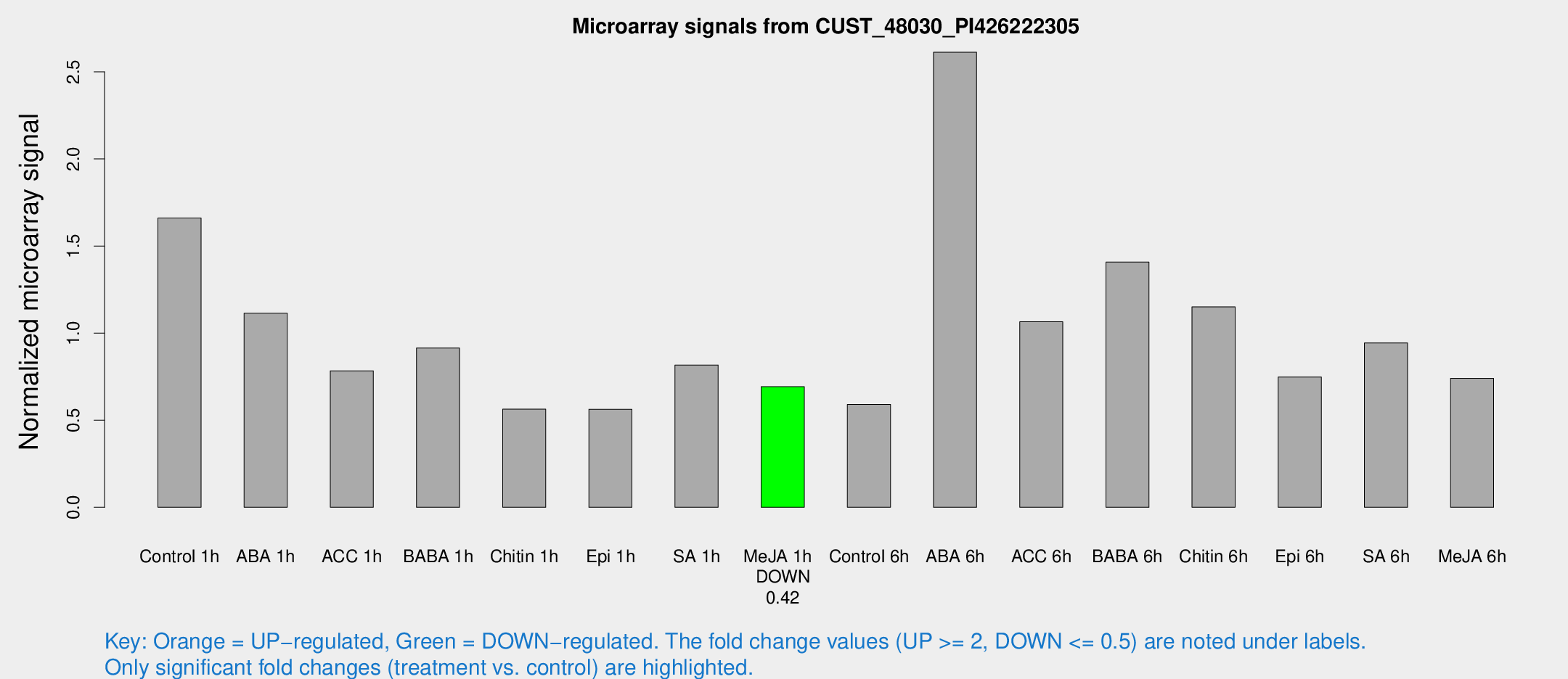
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 79.0008 | 8.72534 | 1.66051 | 0.114089 |
| ABA 1h | 47.4396 | 7.3757 | 1.11429 | 0.168919 |
| ACC 1h | 42.8708 | 13.1251 | 0.783739 | 0.254471 |
| BABA 1h | 42.5338 | 6.96081 | 0.914338 | 0.166487 |
| Chitin 1h | 31.4459 | 14.7938 | 0.563203 | 0.375827 |
| Epi 1h | 27.1031 | 10.5061 | 0.562715 | 0.253795 |
| SA 1h | 41.3248 | 8.08178 | 0.817083 | 0.193364 |
| Me-JA 1h | 26.7271 | 3.32143 | 0.692673 | 0.0860351 |
| Control 6h | 39.6685 | 22.1981 | 0.590748 | 0.406667 |
| ABA 6h | 130.738 | 8.16688 | 2.61248 | 0.163041 |
| ACC 6h | 65.078 | 19.2205 | 1.06496 | 0.37091 |
| BABA 6h | 76.8445 | 13.9597 | 1.40743 | 0.222174 |
| Chitin 6h | 58.4303 | 7.17678 | 1.15065 | 0.108665 |
| Epi 6h | 40.457 | 5.68366 | 0.74759 | 0.0774454 |
| SA 6h | 45.3902 | 8.08599 | 0.943519 | 0.0914056 |
| Me-JA 6h | 36.4819 | 8.37824 | 0.741042 | 0.194503 |
Source Transcript PGSC0003DMT400021782 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
