Probe CUST_47888_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47888_PI426222305 | JHI_St_60k_v1 | DMT400019426 | GAGGCATTTCATTATCCTGGACAAGTATCTTTGTACTACCAAGAGTTCCCAATAACCTTA |
All Microarray Probes Designed to Gene DMG400007511
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47888_PI426222305 | JHI_St_60k_v1 | DMT400019426 | GAGGCATTTCATTATCCTGGACAAGTATCTTTGTACTACCAAGAGTTCCCAATAACCTTA |
Microarray Signals from CUST_47888_PI426222305
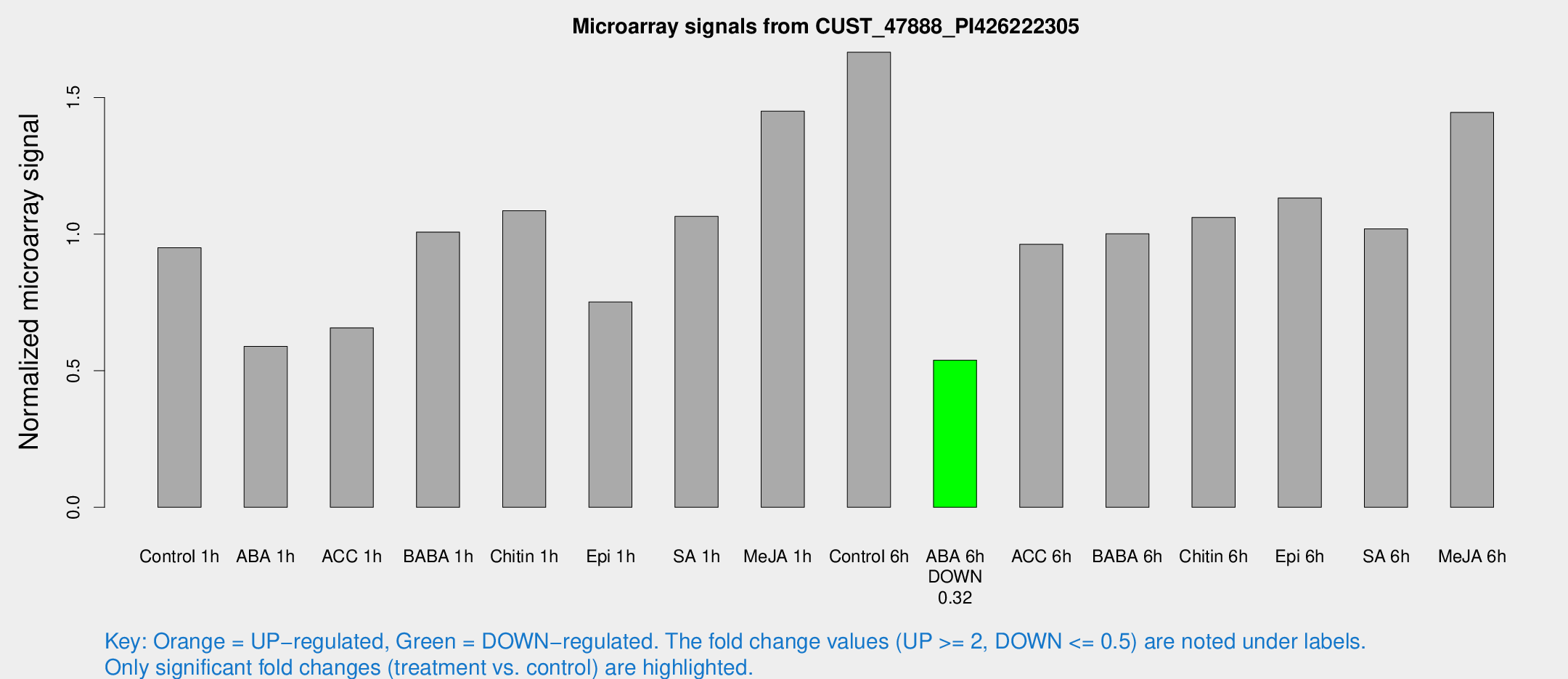
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 335.569 | 69.3973 | 0.950393 | 0.128617 |
| ABA 1h | 177.583 | 11.0282 | 0.589118 | 0.0666372 |
| ACC 1h | 237.126 | 40.9 | 0.657064 | 0.0770037 |
| BABA 1h | 339.293 | 56.1939 | 1.00764 | 0.0865059 |
| Chitin 1h | 338.081 | 45.942 | 1.086 | 0.118767 |
| Epi 1h | 227.566 | 40.9495 | 0.751717 | 0.10938 |
| SA 1h | 377.043 | 47.9149 | 1.06503 | 0.148114 |
| Me-JA 1h | 409.062 | 52.1369 | 1.45017 | 0.0872683 |
| Control 6h | 629.172 | 173.304 | 1.6659 | 0.442266 |
| ABA 6h | 195.298 | 13.3138 | 0.538682 | 0.0323087 |
| ACC 6h | 392.122 | 83.9989 | 0.962649 | 0.0832833 |
| BABA 6h | 384.498 | 39.7605 | 1.00142 | 0.0877099 |
| Chitin 6h | 386.579 | 33.6405 | 1.06143 | 0.0632877 |
| Epi 6h | 442.584 | 66.0507 | 1.13226 | 0.19676 |
| SA 6h | 390.892 | 118.521 | 1.01953 | 0.298363 |
| Me-JA 6h | 495.08 | 55.2227 | 1.44555 | 0.112404 |
Source Transcript PGSC0003DMT400019426 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G45650.1 | +2 | 6e-180 | 525 | 290/546 (53%) | nitrate excretion transporter1 | chr3:16759253-16761266 FORWARD LENGTH=558 |
