Probe CUST_47803_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47803_PI426222305 | JHI_St_60k_v1 | DMT400019331 | CTTGTTTGATTTCTTCACAAGAATATCAAGAGATCGTCATCAATGGTGGCTACGATGAAA |
All Microarray Probes Designed to Gene DMG400007472
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47803_PI426222305 | JHI_St_60k_v1 | DMT400019331 | CTTGTTTGATTTCTTCACAAGAATATCAAGAGATCGTCATCAATGGTGGCTACGATGAAA |
Microarray Signals from CUST_47803_PI426222305
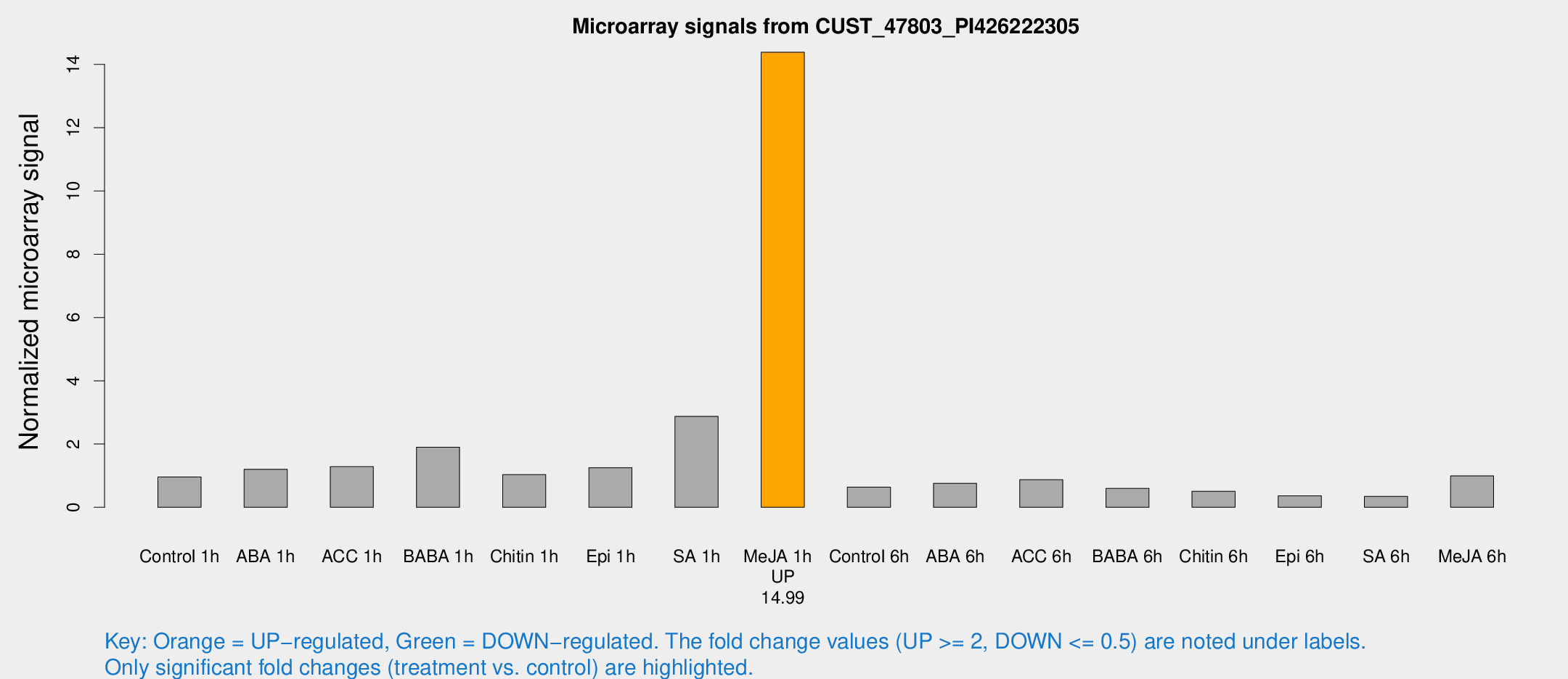
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 19.2152 | 7.61241 | 0.959993 | 0.338726 |
| ABA 1h | 26.9199 | 13.6049 | 1.20245 | 1.30647 |
| ACC 1h | 30.8571 | 15.6706 | 1.28431 | 0.80604 |
| BABA 1h | 31.7377 | 3.81893 | 1.90233 | 0.229772 |
| Chitin 1h | 16.0658 | 3.29684 | 1.03282 | 0.212172 |
| Epi 1h | 20.0643 | 4.73046 | 1.25098 | 0.348597 |
| SA 1h | 59.9044 | 23.1853 | 2.87618 | 1.45716 |
| Me-JA 1h | 205.212 | 21.1764 | 14.3869 | 1.07759 |
| Control 6h | 14.7935 | 8.17201 | 0.636388 | 0.343948 |
| ABA 6h | 17.0127 | 6.33178 | 0.759405 | 0.380128 |
| ACC 6h | 29.3724 | 20.0206 | 0.870918 | 1.18889 |
| BABA 6h | 14.1065 | 6.41763 | 0.597193 | 0.285302 |
| Chitin 6h | 11.0595 | 4.74216 | 0.50843 | 0.235095 |
| Epi 6h | 7.1131 | 3.84213 | 0.363506 | 0.195718 |
| SA 6h | 5.93531 | 3.43975 | 0.346705 | 0.20077 |
| Me-JA 6h | 29.1289 | 20.7122 | 0.992705 | 1.33621 |
Source Transcript PGSC0003DMT400019331 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G24580.1 | +1 | 3e-15 | 70 | 33/71 (46%) | RING/U-box superfamily protein | chr1:8710232-8710573 FORWARD LENGTH=113 |
