Probe CUST_47636_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47636_PI426222305 | JHI_St_60k_v1 | DMT400005182 | GCACCTACCGCTAGACAATTACCACCTTACTAAAATCGTATTCAGAATGTGAAGTTTACA |
All Microarray Probes Designed to Gene DMG400002047
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47636_PI426222305 | JHI_St_60k_v1 | DMT400005182 | GCACCTACCGCTAGACAATTACCACCTTACTAAAATCGTATTCAGAATGTGAAGTTTACA |
Microarray Signals from CUST_47636_PI426222305
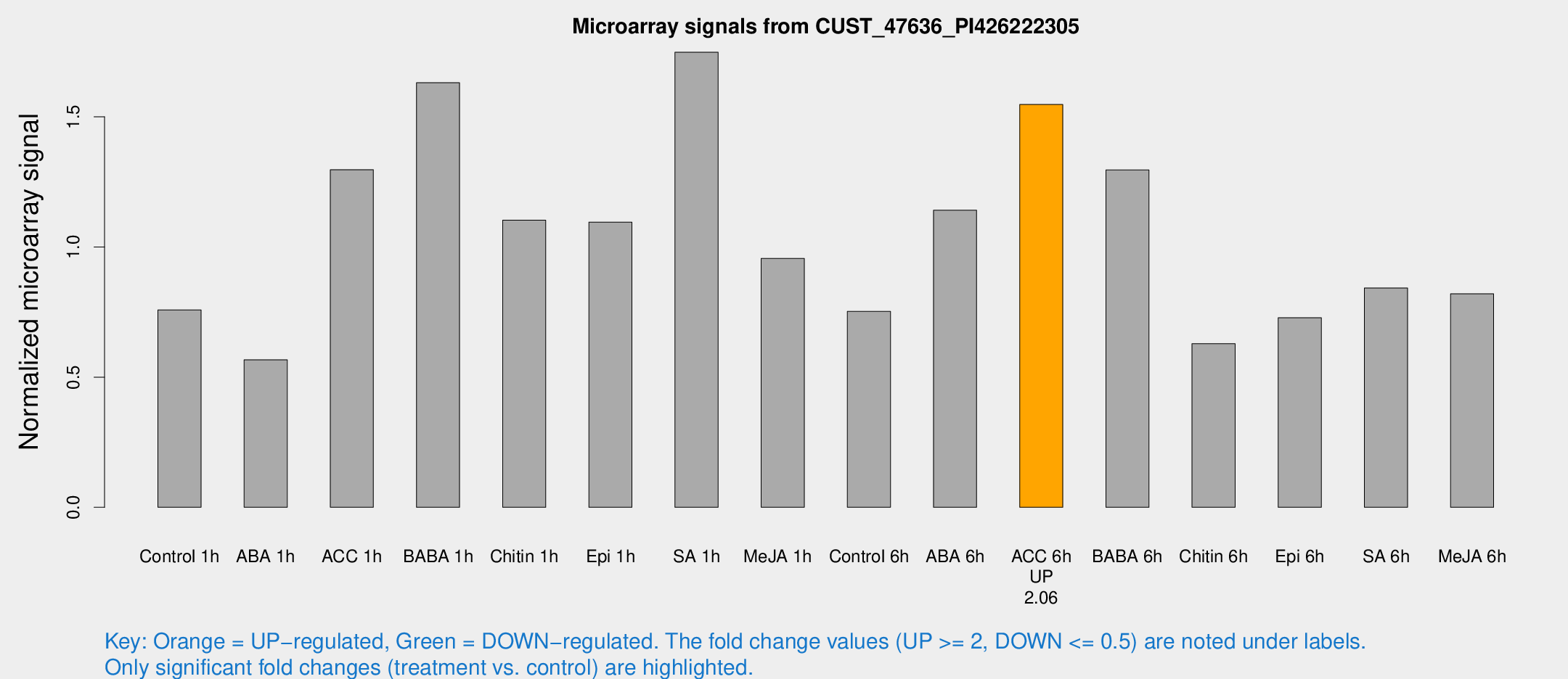
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1434.4 | 188.774 | 0.758377 | 0.080213 |
| ABA 1h | 1024.52 | 271.738 | 0.566573 | 0.165534 |
| ACC 1h | 3007.04 | 1059.1 | 1.29692 | 0.607465 |
| BABA 1h | 3087.2 | 681.217 | 1.63106 | 0.236848 |
| Chitin 1h | 1855.15 | 141.86 | 1.10306 | 0.063724 |
| Epi 1h | 1774.38 | 139.404 | 1.09553 | 0.0832062 |
| SA 1h | 3462.82 | 664.961 | 1.74821 | 0.226544 |
| Me-JA 1h | 1491.22 | 255.337 | 0.956101 | 0.0958906 |
| Control 6h | 1455.8 | 259.318 | 0.752643 | 0.0785883 |
| ABA 6h | 2422.95 | 616.95 | 1.14113 | 0.259401 |
| ACC 6h | 3320.61 | 258.124 | 1.54754 | 0.121354 |
| BABA 6h | 2883.84 | 780.78 | 1.29583 | 0.294201 |
| Chitin 6h | 1243.82 | 71.9434 | 0.628314 | 0.0363377 |
| Epi 6h | 1567.44 | 237.378 | 0.72826 | 0.0757901 |
| SA 6h | 1623.3 | 338.535 | 0.842305 | 0.0999538 |
| Me-JA 6h | 1556.35 | 237.966 | 0.819948 | 0.096215 |
Source Transcript PGSC0003DMT400005182 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G49700.1 | +3 | 1e-42 | 152 | 132/286 (46%) | Predicted AT-hook DNA-binding family protein | chr5:20192599-20193429 FORWARD LENGTH=276 |
