Probe CUST_46932_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46932_PI426222305 | JHI_St_60k_v1 | DMT400048698 | GGTTTCAGGTTTTGGACGTACATATTTTGCCAAGGATGAATCAATTGGATTCTTAATCAA |
All Microarray Probes Designed to Gene DMG400018922
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46932_PI426222305 | JHI_St_60k_v1 | DMT400048698 | GGTTTCAGGTTTTGGACGTACATATTTTGCCAAGGATGAATCAATTGGATTCTTAATCAA |
Microarray Signals from CUST_46932_PI426222305
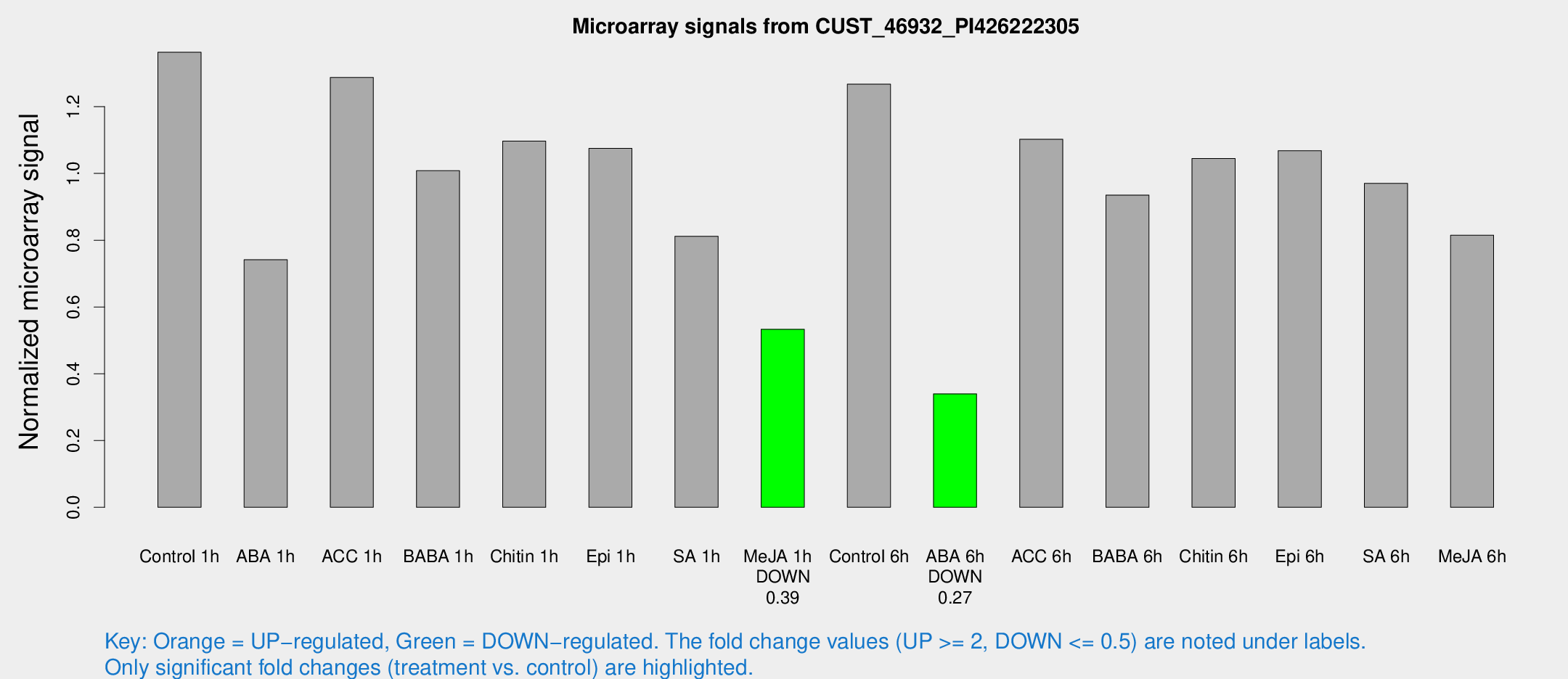
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1650.06 | 142.788 | 1.36311 | 0.078758 |
| ABA 1h | 790.161 | 45.8898 | 0.741454 | 0.0665183 |
| ACC 1h | 1665.29 | 331.78 | 1.28757 | 0.196991 |
| BABA 1h | 1273.3 | 331.069 | 1.00816 | 0.215535 |
| Chitin 1h | 1188.73 | 68.9432 | 1.09688 | 0.0682471 |
| Epi 1h | 1122.53 | 74.1203 | 1.07494 | 0.0800463 |
| SA 1h | 1035.86 | 196.007 | 0.811923 | 0.101158 |
| Me-JA 1h | 532.105 | 68.9823 | 0.53333 | 0.0310384 |
| Control 6h | 1544.97 | 174.272 | 1.26738 | 0.180652 |
| ABA 6h | 436.122 | 29.4117 | 0.339841 | 0.0236272 |
| ACC 6h | 1541.28 | 188.834 | 1.10255 | 0.0637373 |
| BABA 6h | 1275.08 | 153.664 | 0.935216 | 0.104349 |
| Chitin 6h | 1342.82 | 92.7523 | 1.04471 | 0.0604121 |
| Epi 6h | 1448.83 | 83.876 | 1.06809 | 0.0617549 |
| SA 6h | 1291.03 | 373.409 | 0.970175 | 0.244822 |
| Me-JA 6h | 1010.27 | 181.778 | 0.81508 | 0.0877343 |
Source Transcript PGSC0003DMT400048698 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G31350.1 | +2 | 8e-97 | 301 | 192/379 (51%) | KAR-UP F-box 1 | chr1:11221519-11222706 REVERSE LENGTH=395 |
