Probe CUST_46815_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46815_PI426222305 | JHI_St_60k_v1 | DMT400001874 | CATGCAGTTAGGTTGCAGAAATTCTGCGGATTTCAAAAGGTTATGTTAGCTGGTAGGTTA |
All Microarray Probes Designed to Gene DMG400000706
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46796_PI426222305 | JHI_St_60k_v1 | DMT400001872 | CATGCAGTTAGGTTGCAGAAATTCTGCGGATTTCAAAAGGTTATGTTAGCTGGTAGGTTA |
| CUST_46815_PI426222305 | JHI_St_60k_v1 | DMT400001874 | CATGCAGTTAGGTTGCAGAAATTCTGCGGATTTCAAAAGGTTATGTTAGCTGGTAGGTTA |
| CUST_46800_PI426222305 | JHI_St_60k_v1 | DMT400001873 | GGTTATAGGTCTAAATGAATGTAGCACATGATTTATGTTTCTAGTGAACATTCACCCCCC |
Microarray Signals from CUST_46815_PI426222305
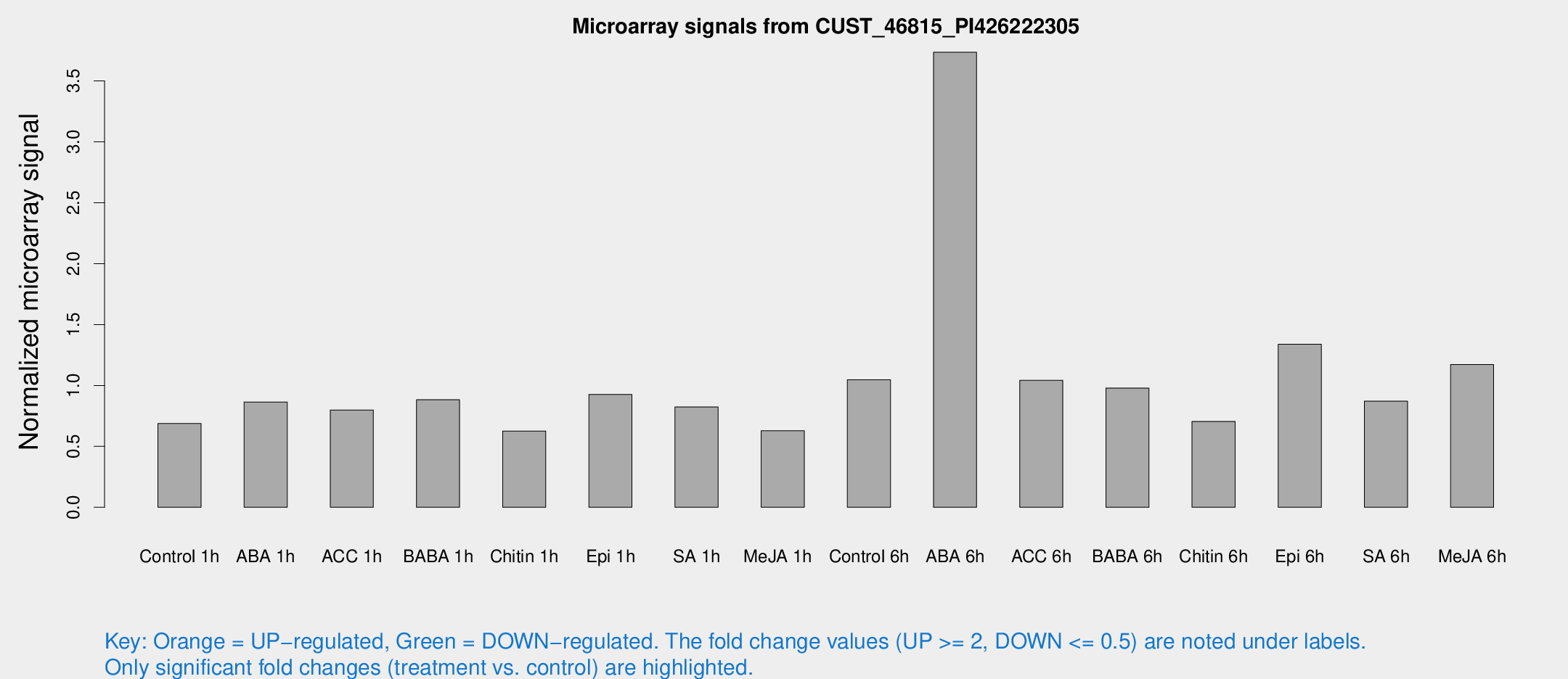
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 15.6867 | 4.61392 | 0.687794 | 0.292076 |
| ABA 1h | 20.4726 | 11.2066 | 0.8646 | 0.454609 |
| ACC 1h | 17.8784 | 4.51616 | 0.798801 | 0.270333 |
| BABA 1h | 19.7279 | 6.99054 | 0.883021 | 0.273839 |
| Chitin 1h | 11.8947 | 3.71009 | 0.626222 | 0.228589 |
| Epi 1h | 18.1128 | 5.14264 | 0.926611 | 0.361483 |
| SA 1h | 20.2739 | 7.60615 | 0.82359 | 0.396888 |
| Me-JA 1h | 11.9985 | 4.7614 | 0.62833 | 0.259481 |
| Control 6h | 23.9073 | 7.02216 | 1.04683 | 0.312643 |
| ABA 6h | 83.2819 | 16.9572 | 3.73538 | 0.529437 |
| ACC 6h | 29.2571 | 10.3532 | 1.04219 | 0.46846 |
| BABA 6h | 24.6065 | 8.47349 | 0.978765 | 0.296931 |
| Chitin 6h | 15.3606 | 4.41894 | 0.705205 | 0.210705 |
| Epi 6h | 30.6089 | 5.10863 | 1.33846 | 0.227167 |
| SA 6h | 20.5678 | 6.92903 | 0.870639 | 0.313276 |
| Me-JA 6h | 28.8698 | 11.5777 | 1.17159 | 0.570826 |
Source Transcript PGSC0003DMT400001874 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
