Probe CUST_46806_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46806_PI426222305 | JHI_St_60k_v1 | DMT400001937 | TCTTATGCCTTGCTGACATGCATGATAGCTCATGTTTCTGGTCTAGTTCCTGGTGATTTC |
All Microarray Probes Designed to Gene DMG400000736
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46799_PI426222305 | JHI_St_60k_v1 | DMT400001938 | ACTATTCACCTCATGATGTTTGCTCTACTTCTCTTTACAGGTCTAGTTCCTGGTGATTTC |
| CUST_46806_PI426222305 | JHI_St_60k_v1 | DMT400001937 | TCTTATGCCTTGCTGACATGCATGATAGCTCATGTTTCTGGTCTAGTTCCTGGTGATTTC |
Microarray Signals from CUST_46806_PI426222305
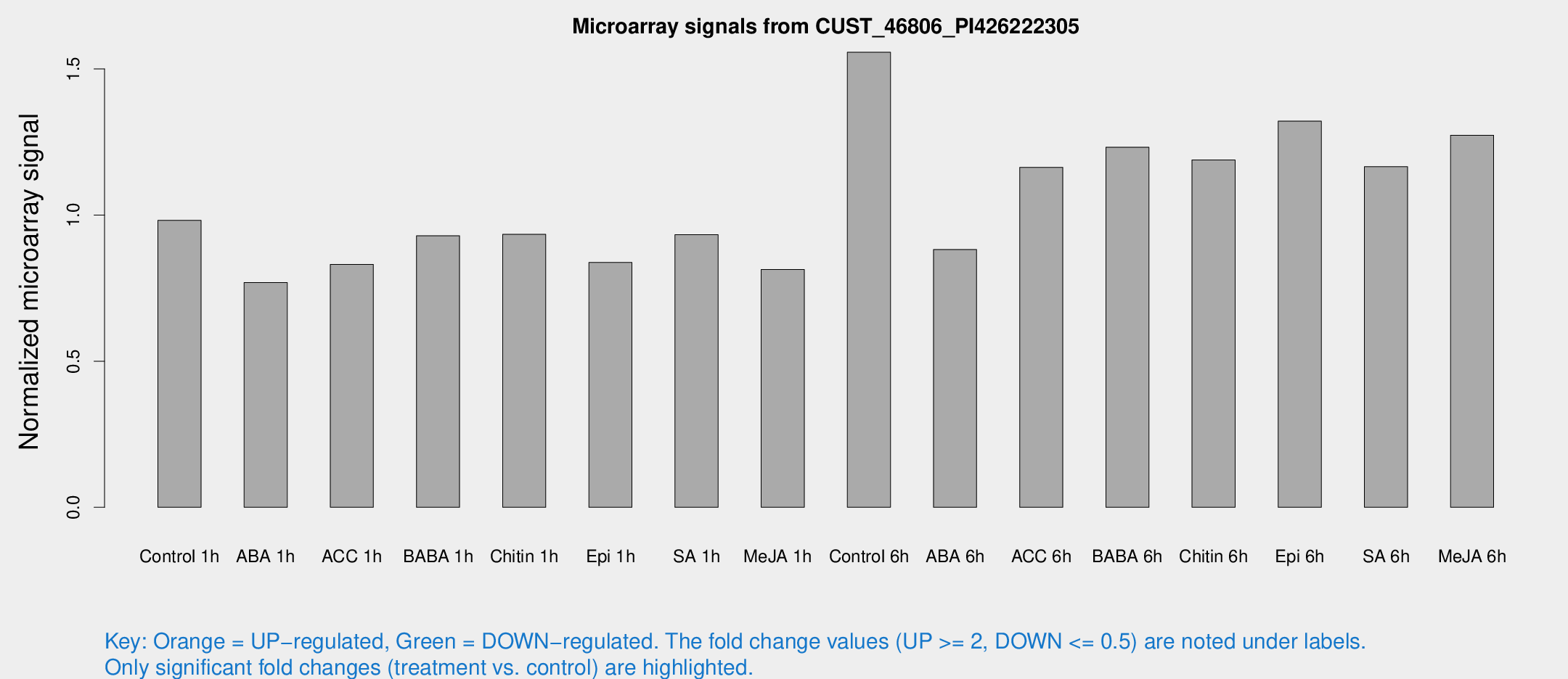
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 914.602 | 85.0009 | 0.982064 | 0.0568401 |
| ABA 1h | 632.552 | 54.7226 | 0.76906 | 0.0446156 |
| ACC 1h | 842.632 | 196.865 | 0.831069 | 0.164442 |
| BABA 1h | 850.392 | 137.565 | 0.929241 | 0.0728056 |
| Chitin 1h | 784.163 | 80.9486 | 0.934143 | 0.0541096 |
| Epi 1h | 676.887 | 71.1207 | 0.837755 | 0.0703948 |
| SA 1h | 889.797 | 66.3879 | 0.932889 | 0.054 |
| Me-JA 1h | 615.758 | 40.5452 | 0.813623 | 0.0472398 |
| Control 6h | 1564.29 | 404.719 | 1.55729 | 0.346255 |
| ABA 6h | 869.326 | 54.8111 | 0.882251 | 0.0511056 |
| ACC 6h | 1262.64 | 205.164 | 1.1634 | 0.0672925 |
| BABA 6h | 1275.92 | 73.9596 | 1.23212 | 0.0714755 |
| Chitin 6h | 1177.88 | 107.873 | 1.18838 | 0.0751941 |
| Epi 6h | 1399.69 | 187.089 | 1.32168 | 0.156026 |
| SA 6h | 1134.19 | 268.998 | 1.16557 | 0.178209 |
| Me-JA 6h | 1215.95 | 229.425 | 1.27328 | 0.145447 |
Source Transcript PGSC0003DMT400001937 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G16370.1 | +1 | 0.0 | 818 | 394/508 (78%) | thymidylate synthase 1 | chr2:7082038-7084333 REVERSE LENGTH=519 |
