Probe CUST_46461_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46461_PI426222305 | JHI_St_60k_v1 | DMT400016330 | GATTATGTGATTTTGGAAATGTTTTGTGTAATGTTTTCGTGGGATTGGGATATGCAAGTG |
All Microarray Probes Designed to Gene DMG400006383
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46461_PI426222305 | JHI_St_60k_v1 | DMT400016330 | GATTATGTGATTTTGGAAATGTTTTGTGTAATGTTTTCGTGGGATTGGGATATGCAAGTG |
Microarray Signals from CUST_46461_PI426222305
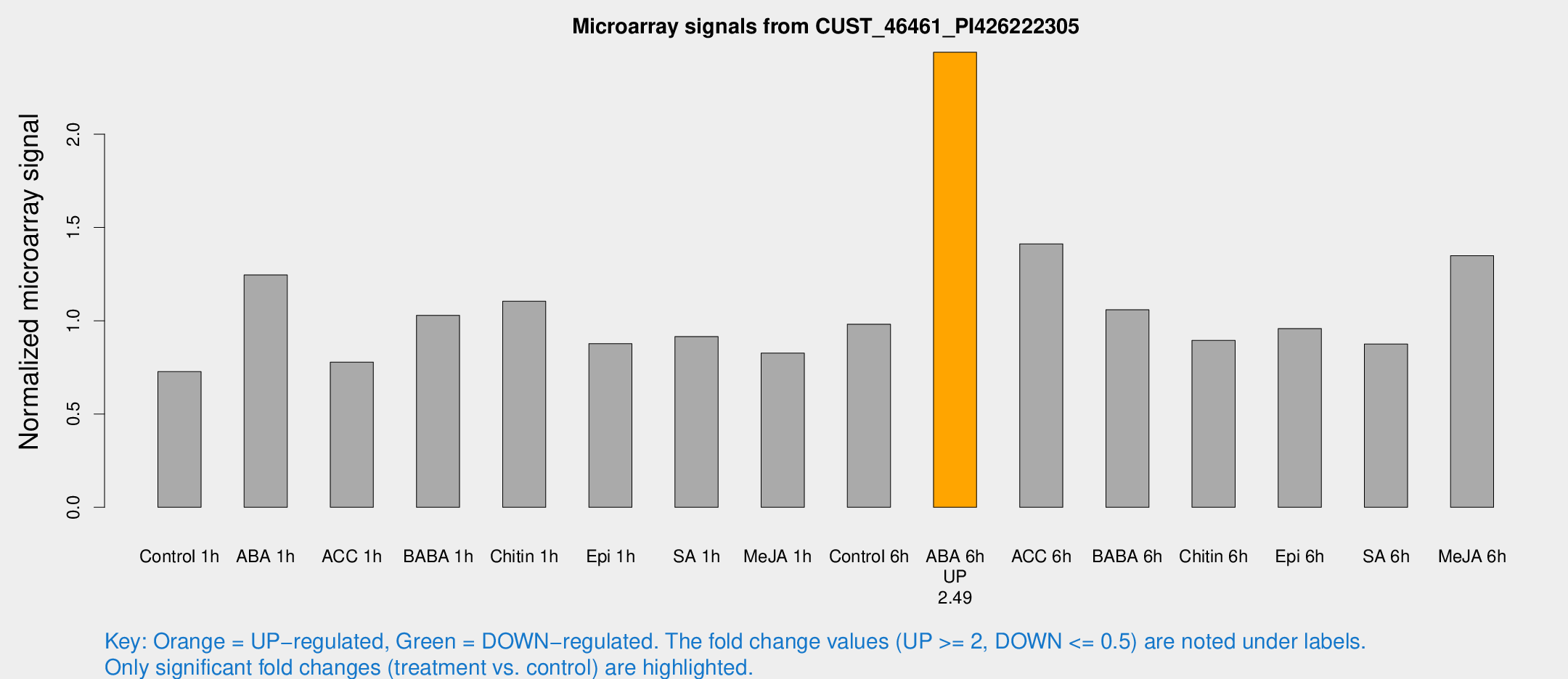
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6307.87 | 365.401 | 0.726965 | 0.0630787 |
| ABA 1h | 9731.78 | 1350.67 | 1.24527 | 0.136388 |
| ACC 1h | 7504.58 | 1909.64 | 0.777993 | 0.184765 |
| BABA 1h | 9047.1 | 1907.58 | 1.02891 | 0.14253 |
| Chitin 1h | 8668.49 | 856.171 | 1.10458 | 0.0637741 |
| Epi 1h | 6604.54 | 525.214 | 0.876981 | 0.0721088 |
| SA 1h | 8417.4 | 1600.72 | 0.91535 | 0.114516 |
| Me-JA 1h | 5905.9 | 651.02 | 0.826157 | 0.0477 |
| Control 6h | 8646.29 | 1052.9 | 0.981478 | 0.0566667 |
| ABA 6h | 22944.9 | 3428.86 | 2.43914 | 0.239355 |
| ACC 6h | 14028.9 | 811.214 | 1.41209 | 0.140476 |
| BABA 6h | 10378.3 | 1186.47 | 1.05867 | 0.0805258 |
| Chitin 6h | 8238.96 | 476.152 | 0.894171 | 0.0516262 |
| Epi 6h | 9422.5 | 851.798 | 0.95819 | 0.0553222 |
| SA 6h | 8431.54 | 2445.48 | 0.874788 | 0.232983 |
| Me-JA 6h | 11760.5 | 1273.12 | 1.34883 | 0.0778753 |
Source Transcript PGSC0003DMT400016330 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G64430.1 | +2 | 5e-67 | 231 | 191/471 (41%) | Octicosapeptide/Phox/Bem1p family protein | chr5:25762540-25764081 REVERSE LENGTH=513 |
