Probe CUST_46456_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46456_PI426222305 | JHI_St_60k_v1 | DMT400016361 | CAAGTCTCTGTTCTCTACTTCATTGACTGATCTACTGAATTTAGATCATGTAGCTTGTTA |
All Microarray Probes Designed to Gene DMG400006395
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46456_PI426222305 | JHI_St_60k_v1 | DMT400016361 | CAAGTCTCTGTTCTCTACTTCATTGACTGATCTACTGAATTTAGATCATGTAGCTTGTTA |
Microarray Signals from CUST_46456_PI426222305
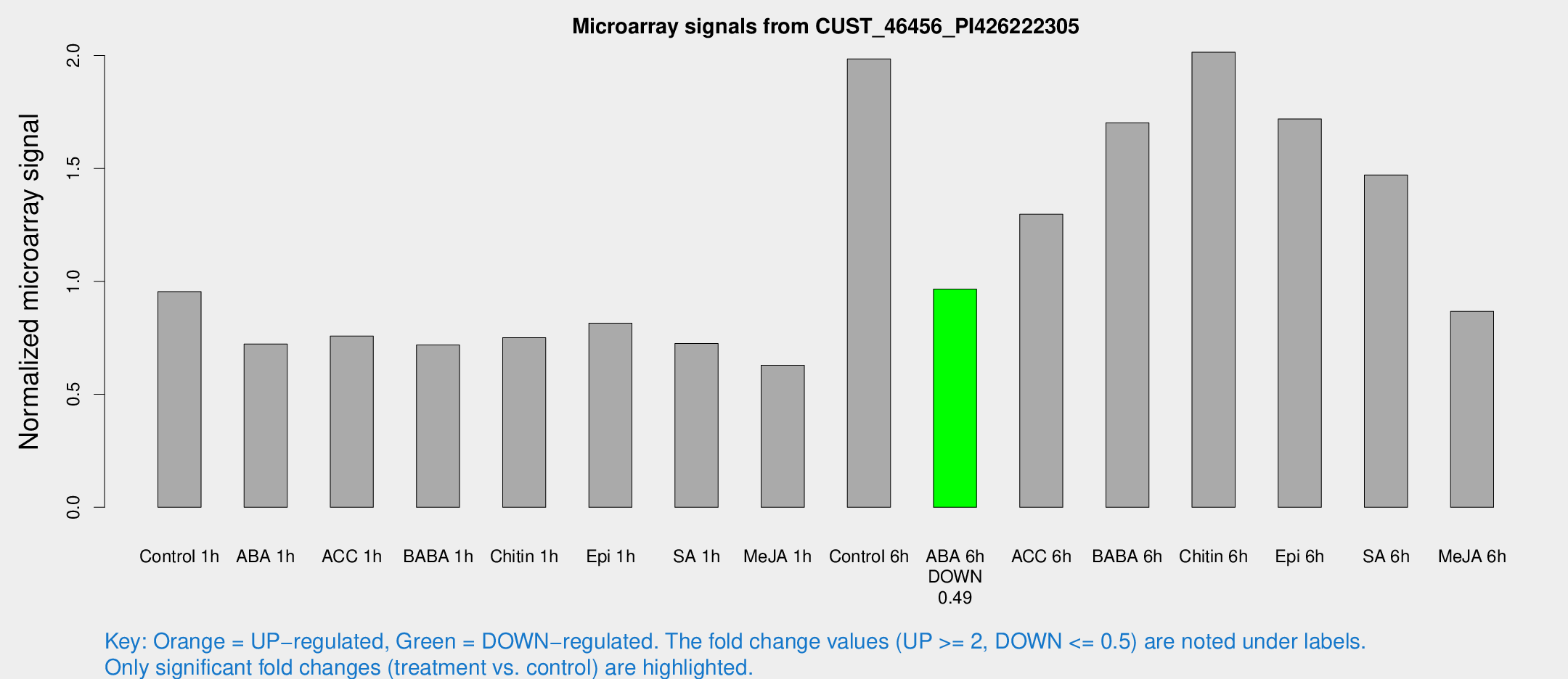
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 642.65 | 107.144 | 0.954381 | 0.104823 |
| ABA 1h | 423.112 | 46.4952 | 0.723002 | 0.0593218 |
| ACC 1h | 540.143 | 126.961 | 0.757647 | 0.135659 |
| BABA 1h | 460.549 | 57.3009 | 0.718571 | 0.0418326 |
| Chitin 1h | 453.581 | 76.9488 | 0.750922 | 0.151785 |
| Epi 1h | 464.504 | 36.9797 | 0.815382 | 0.053622 |
| SA 1h | 495.35 | 63.9399 | 0.724774 | 0.124912 |
| Me-JA 1h | 339.987 | 36.7862 | 0.629119 | 0.0770855 |
| Control 6h | 1379.87 | 310.546 | 1.98484 | 0.341575 |
| ABA 6h | 673.289 | 39.1338 | 0.965889 | 0.0559806 |
| ACC 6h | 1004.87 | 181.931 | 1.29755 | 0.075087 |
| BABA 6h | 1305.84 | 297.577 | 1.70193 | 0.364632 |
| Chitin 6h | 1409.58 | 106.393 | 2.01432 | 0.116414 |
| Epi 6h | 1290.74 | 165.946 | 1.71891 | 0.365676 |
| SA 6h | 956.553 | 68.4893 | 1.47112 | 0.0851104 |
| Me-JA 6h | 647.167 | 204.33 | 0.867231 | 0.294241 |
Source Transcript PGSC0003DMT400016361 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G12770.1 | +3 | 1e-146 | 453 | 229/671 (34%) | mitochondrial editing factor 22 | chr3:4057027-4059193 REVERSE LENGTH=694 |
