Probe CUST_4641_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4641_PI426222305 | JHI_St_60k_v1 | DMT400070461 | GTAAAGTATTATAAAGAAAATGCTGGTGCTGATGAATCTGTGTTGTTGTATTGGAGTGCT |
All Microarray Probes Designed to Gene DMG401027394
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4641_PI426222305 | JHI_St_60k_v1 | DMT400070461 | GTAAAGTATTATAAAGAAAATGCTGGTGCTGATGAATCTGTGTTGTTGTATTGGAGTGCT |
Microarray Signals from CUST_4641_PI426222305
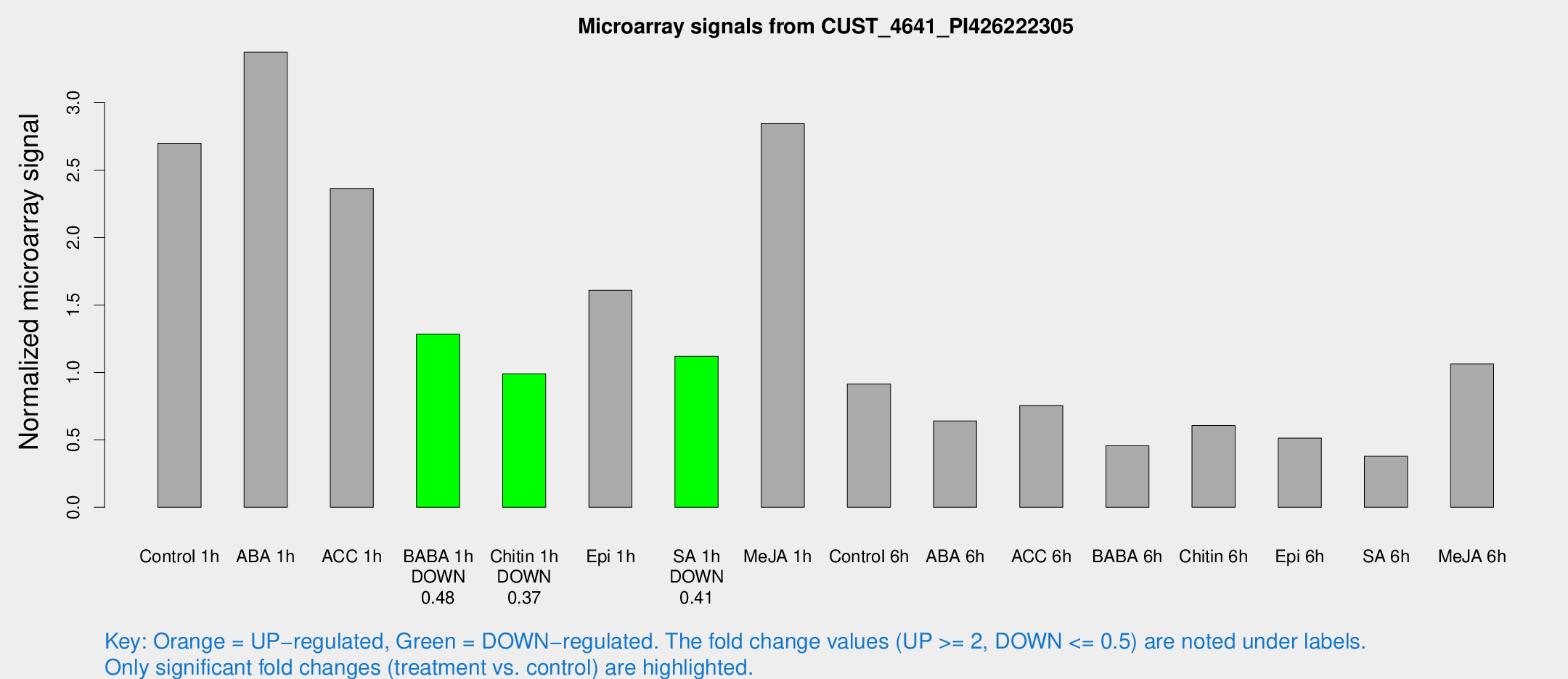
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1021.02 | 158.669 | 2.69855 | 0.295686 |
| ABA 1h | 1132.32 | 187.259 | 3.3737 | 0.562073 |
| ACC 1h | 919.058 | 142.656 | 2.36456 | 0.273459 |
| BABA 1h | 478.927 | 96.2187 | 1.28393 | 0.158863 |
| Chitin 1h | 335.234 | 46.7135 | 0.989955 | 0.153186 |
| Epi 1h | 520.764 | 54.7466 | 1.60837 | 0.167194 |
| SA 1h | 427.901 | 33.6825 | 1.11926 | 0.0766973 |
| Me-JA 1h | 881.981 | 136.133 | 2.84395 | 0.336399 |
| Control 6h | 375.64 | 109.227 | 0.914395 | 0.242636 |
| ABA 6h | 256.003 | 32.0442 | 0.641029 | 0.0741505 |
| ACC 6h | 338.949 | 85.3608 | 0.754088 | 0.0734253 |
| BABA 6h | 192.253 | 26.0958 | 0.455979 | 0.069524 |
| Chitin 6h | 242.06 | 28.3867 | 0.606317 | 0.0529861 |
| Epi 6h | 219.461 | 37.3497 | 0.51218 | 0.0975379 |
| SA 6h | 163.708 | 55.2032 | 0.377914 | 0.136083 |
| Me-JA 6h | 393.342 | 27.7502 | 1.06287 | 0.15843 |
Source Transcript PGSC0003DMT400070461 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G55290.1 | +2 | 4e-38 | 137 | 69/171 (40%) | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | chr1:20626208-20627397 REVERSE LENGTH=361 |
